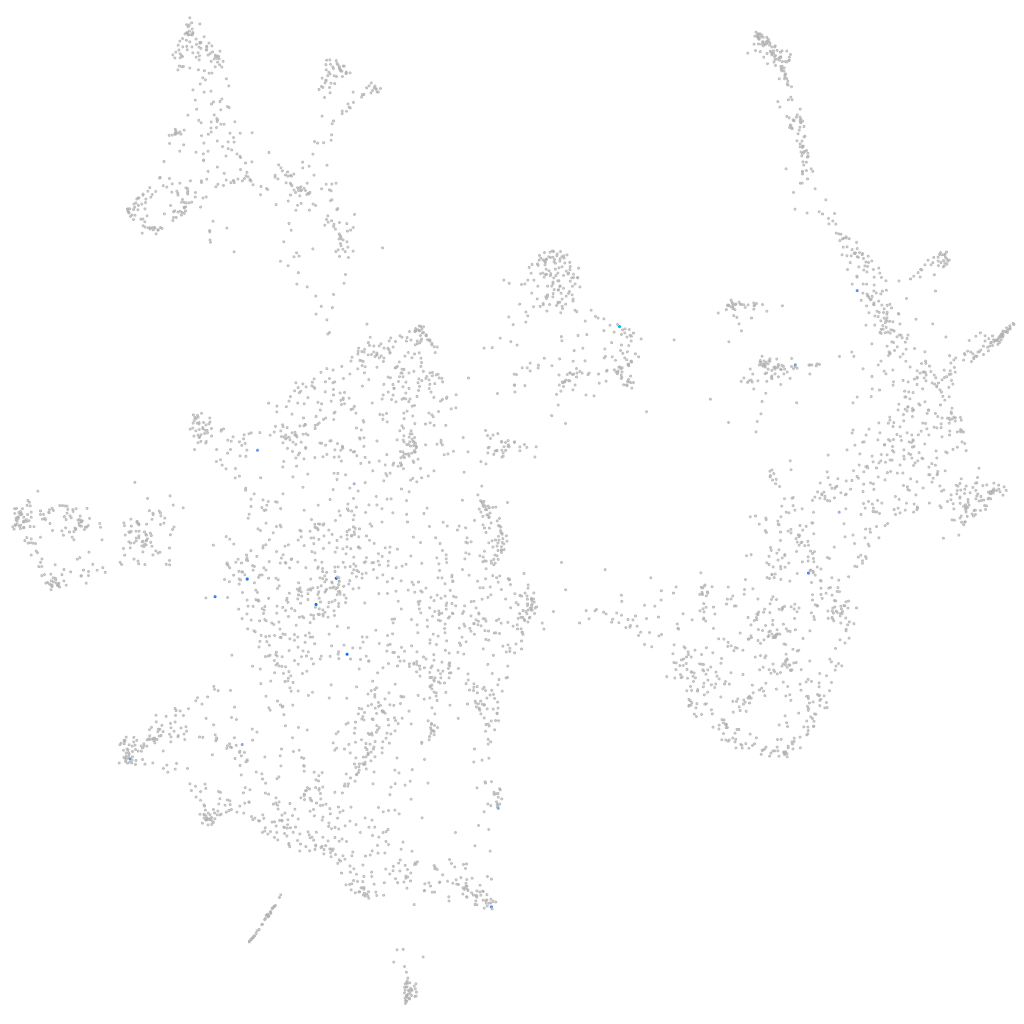
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia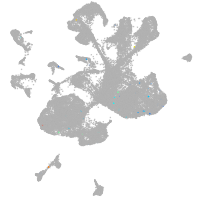 |
eye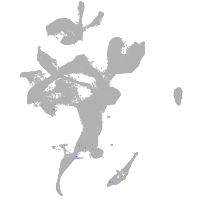 |
taste / olfactory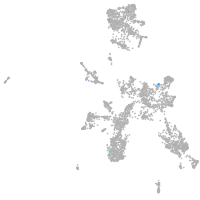 |
otic / lateral line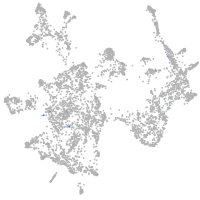 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| or106-8 | 0.522 | actc1b | -0.034 |
| foxl1 | 0.345 | pvalb1 | -0.029 |
| AL928977.1 | 0.287 | calm1b | -0.027 |
| RF00415 | 0.266 | gapdhs | -0.027 |
| XLOC-008016 | 0.261 | rnasekb | -0.026 |
| CR925804.1 | 0.256 | atp6v0ca | -0.026 |
| si:ch73-380l3.4 | 0.239 | mylpfa | -0.026 |
| mir199-2 | 0.236 | ccni | -0.026 |
| lrp13 | 0.223 | eno1a | -0.025 |
| LOC101883911 | 0.217 | mt-nd3 | -0.025 |
| lratb.1 | 0.212 | vamp8 | -0.025 |
| LOC100331306 | 0.206 | cdc34a | -0.024 |
| LOC103910390 | 0.206 | tnni2a.4 | -0.024 |
| fybb | 0.205 | pvalb2 | -0.023 |
| BX649499.2 | 0.202 | vamp2 | -0.022 |
| limd1b | 0.194 | foxg1a | -0.022 |
| foxf2a | 0.191 | ankrd13c | -0.022 |
| CU855900.1 | 0.190 | atp6v1e1b | -0.022 |
| hoxb4a | 0.188 | pvalb8 | -0.021 |
| prrx1a | 0.185 | cd37 | -0.021 |
| twist1b | 0.182 | atf6 | -0.021 |
| rflna | 0.174 | myhz1.1 | -0.021 |
| CR759907.1 | 0.171 | soul4 | -0.021 |
| LOC101883884 | 0.171 | igfbp5b | -0.021 |
| si:dkey-191j3.2 | 0.162 | rtn1a | -0.021 |
| fam129ab | 0.162 | ccdc9 | -0.021 |
| hao2 | 0.160 | wdfy1 | -0.021 |
| CR352243.2 | 0.153 | arfip1 | -0.021 |
| XLOC-042292 | 0.153 | copz2 | -0.021 |
| zgc:112970 | 0.151 | wu:fi46h04 | -0.021 |
| XLOC-018977 | 0.147 | creb3l3l | -0.021 |
| moxd1 | 0.143 | calml4a | -0.021 |
| grnas | 0.141 | pcsk5a | -0.021 |
| hsd17b2 | 0.140 | pdia4 | -0.020 |
| yrk | 0.140 | btc | -0.020 |