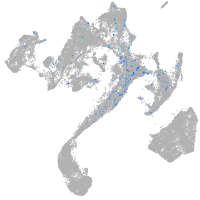
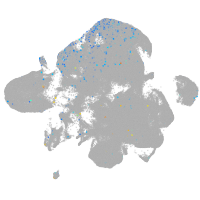
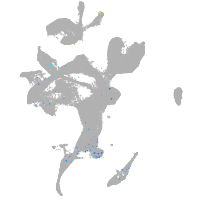
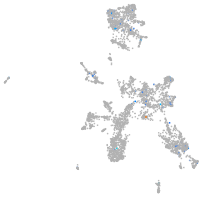
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CABZ01055707.1 | 0.105 | ptmab | -0.052 |
| bmpr1ba | 0.080 | ckbb | -0.046 |
| LOC100536949 | 0.078 | ptmaa | -0.045 |
| cnr2 | 0.078 | gpm6aa | -0.043 |
| pimr80 | 0.078 | hmgb1a | -0.039 |
| CABZ01075068.1 | 0.077 | hmgn6 | -0.037 |
| si:dkey-217m5.8 | 0.077 | gpm6ab | -0.036 |
| eif4ebp3l | 0.074 | si:ch73-1a9.3 | -0.035 |
| CU929070.1 | 0.072 | pvalb1 | -0.034 |
| si:dkey-102m7.3 | 0.068 | pvalb2 | -0.033 |
| snu13b | 0.068 | scrt2 | -0.032 |
| paics | 0.068 | neurod4 | -0.032 |
| npm1a | 0.067 | nova2 | -0.032 |
| vcana | 0.067 | rtn1a | -0.032 |
| cad | 0.067 | btg1 | -0.031 |
| gart | 0.066 | tuba1c | -0.031 |
| zbtb16a | 0.066 | epb41a | -0.030 |
| nip7 | 0.066 | mpp6b | -0.030 |
| pprc1 | 0.065 | cspg5a | -0.029 |
| zfp36l1a | 0.064 | vsx1 | -0.029 |
| nop58 | 0.064 | ndrg4 | -0.029 |
| nifk | 0.063 | mylpfa | -0.029 |
| chrna9 | 0.063 | marcksl1b | -0.028 |
| nhp2 | 0.063 | chd4a | -0.028 |
| prps1a | 0.063 | tuba1a | -0.028 |
| mrto4 | 0.062 | actc1b | -0.027 |
| plxdc1 | 0.062 | calb2b | -0.027 |
| pwp2h | 0.062 | ubc | -0.027 |
| adi1 | 0.061 | gng13b | -0.026 |
| ftsj3 | 0.060 | snap25b | -0.026 |
| twistnb | 0.060 | myt1b | -0.026 |
| id1 | 0.060 | ccng2 | -0.026 |
| bxdc2 | 0.060 | gnb3a | -0.026 |
| thyn1 | 0.059 | nsg2 | -0.025 |
| selenoh | 0.059 | tp53inp2 | -0.025 |