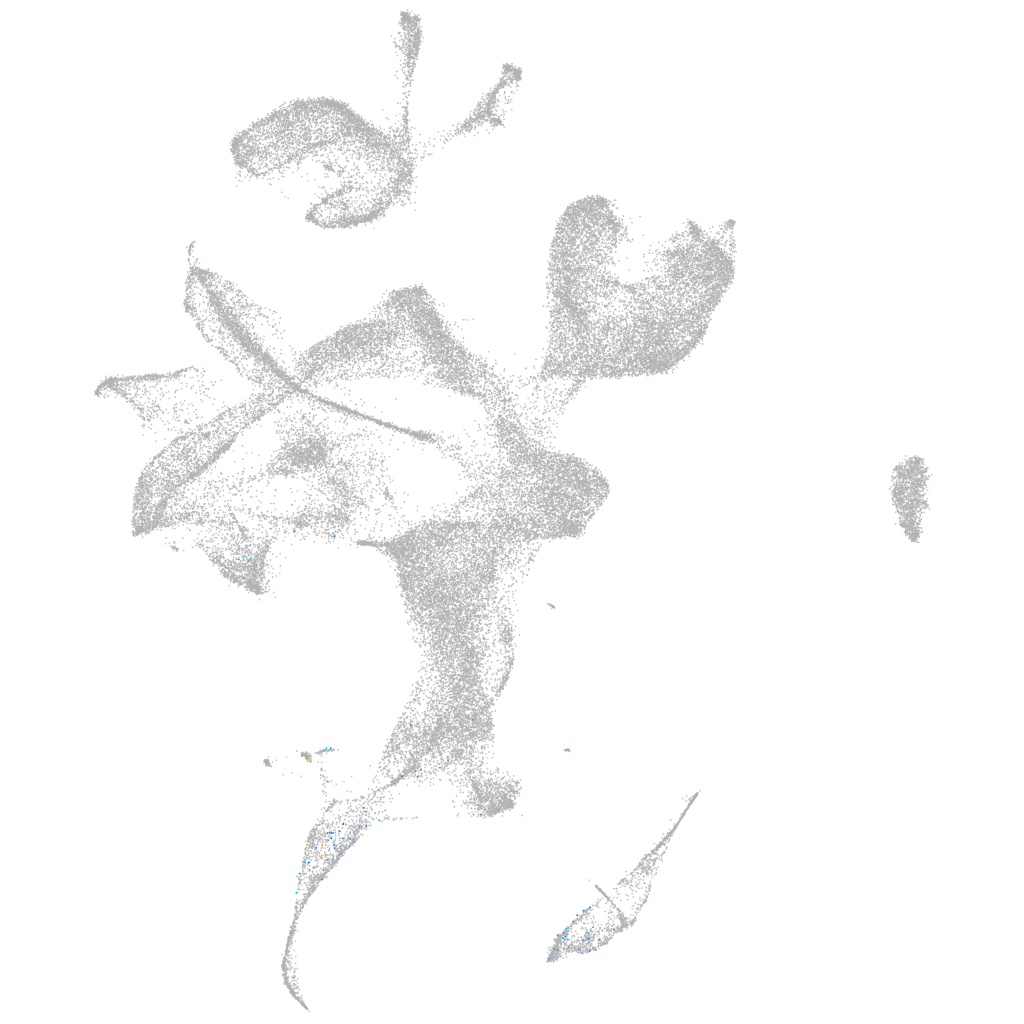
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CABZ01109081.1 | 0.213 | ptmab | -0.064 |
| LOC101884216 | 0.160 | ptmaa | -0.046 |
| si:rp71-45k5.2 | 0.122 | si:ch73-1a9.3 | -0.040 |
| slc3a2a | 0.114 | tuba1c | -0.039 |
| sytl2b | 0.112 | hmgb1a | -0.038 |
| tmem88b | 0.111 | hmgn6 | -0.038 |
| col4a6 | 0.110 | gpm6aa | -0.037 |
| noto | 0.106 | si:ch211-137a8.4 | -0.035 |
| col4a5 | 0.104 | ckbb | -0.034 |
| tas1r1 | 0.103 | anp32e | -0.031 |
| fabp11a | 0.101 | h2afva | -0.031 |
| cldn19 | 0.100 | tuba1a | -0.030 |
| slc6a2 | 0.100 | foxg1b | -0.030 |
| kcnj1a.2 | 0.099 | cadm3 | -0.027 |
| BX927218.2 | 0.096 | gpm6ab | -0.027 |
| emx1 | 0.095 | rorb | -0.026 |
| qdpra | 0.095 | si:ch1073-429i10.3.1 | -0.026 |
| serpinh1b | 0.094 | nova2 | -0.026 |
| wls | 0.094 | fabp7a | -0.026 |
| tyrp1a | 0.094 | hmgb1b | -0.025 |
| tspan36 | 0.093 | hnrnpa0a | -0.025 |
| CR376825.1 | 0.092 | marcksb | -0.025 |
| igfbp7 | 0.092 | snap25b | -0.024 |
| hspg2 | 0.091 | neurod4 | -0.024 |
| fgfr3 | 0.091 | epb41a | -0.023 |
| zgc:110591 | 0.091 | marcksl1b | -0.023 |
| CABZ01074363.1 | 0.090 | si:dkey-276j7.1 | -0.023 |
| rab38 | 0.090 | stmn1b | -0.023 |
| slc45a2 | 0.090 | hmgn2 | -0.023 |
| asip2b | 0.090 | crx | -0.022 |
| cx43 | 0.089 | atp6v0cb | -0.022 |
| ace | 0.089 | ubc | -0.022 |
| pmelb | 0.089 | cspg5a | -0.022 |
| pmela | 0.088 | ndrg4 | -0.022 |
| dct | 0.087 | sypb | -0.022 |