Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 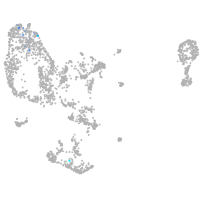 |
neural |
spinal cord / glia |
eye |
taste / olfactory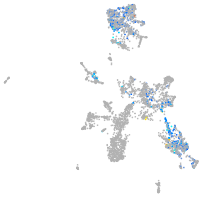 |
otic / lateral line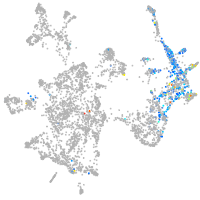 |
epidermis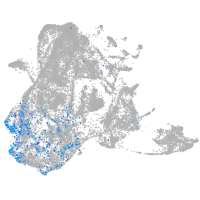 |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| srpx2 | 0.370 | and2 | -0.145 |
| col14a1a | 0.369 | rbp4 | -0.124 |
| has3 | 0.333 | nme2b.1 | -0.097 |
| dlx4b | 0.308 | CABZ01074130.1 | -0.093 |
| vcana | 0.292 | si:dkey-33c14.3 | -0.092 |
| itih5 | 0.291 | aldob | -0.088 |
| sp6 | 0.290 | gapdhs | -0.088 |
| vox | 0.290 | col1a1b | -0.086 |
| gmnn | 0.284 | marcksl1a | -0.085 |
| jag2b | 0.284 | gstp1 | -0.085 |
| slc7a11 | 0.279 | col1a2 | -0.084 |
| nid2a | 0.268 | col5a1 | -0.081 |
| rspo2 | 0.268 | fxyd6l | -0.080 |
| megf6a | 0.265 | pvalb1 | -0.080 |
| tcf7 | 0.265 | wu:fa03e10 | -0.078 |
| hey2 | 0.263 | lrrc17 | -0.076 |
| samd10a | 0.259 | ahcy | -0.076 |
| emid1 | 0.252 | cxl34b.11 | -0.076 |
| plekhh1 | 0.250 | bhmt | -0.075 |
| lamb1a | 0.249 | c1qtnf5 | -0.074 |
| cyp1a | 0.248 | epgn | -0.073 |
| kctd12.2 | 0.247 | gstt1a | -0.072 |
| frem3 | 0.240 | eef1da | -0.072 |
| XLOC-042222 | 0.236 | krtt1c19e | -0.071 |
| alpi.1 | 0.234 | dcn | -0.071 |
| si:ch211-267e7.3 | 0.233 | pvalb2 | -0.070 |
| aplnra | 0.229 | rplp2 | -0.070 |
| fibinb | 0.227 | krt91 | -0.070 |
| cxcr4a | 0.226 | krt5 | -0.070 |
| si:ch211-194g2.4 | 0.226 | cyr61 | -0.070 |
| alcamb | 0.224 | cd82a | -0.069 |
| dlx5a | 0.224 | eif4ebp3 | -0.069 |
| rgs3a | 0.222 | hgd | -0.065 |
| fermt1 | 0.220 | crabp2a | -0.065 |
| apoc1 | 0.218 | pnp5a | -0.064 |




