si:ch211-267e7.3
ZFIN 
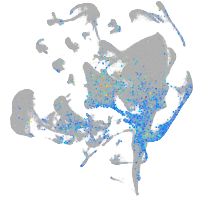
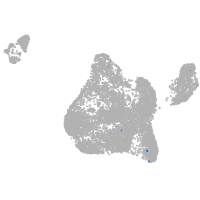
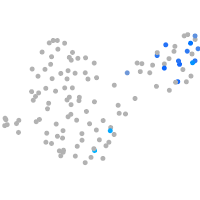
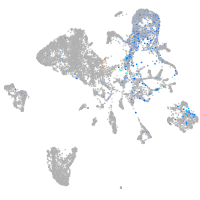
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| nid2a | 0.172 | tuba1c | -0.076 |
| tpm4a | 0.154 | elavl3 | -0.070 |
| mfap2 | 0.148 | gpm6aa | -0.070 |
| fn1a | 0.144 | rtn1a | -0.069 |
| fkbp9 | 0.143 | ckbb | -0.066 |
| lamb1a | 0.141 | nova2 | -0.066 |
| fbn2b | 0.137 | stmn1b | -0.065 |
| si:dkey-261h17.1 | 0.136 | tmsb | -0.061 |
| fkbp7 | 0.135 | gpm6ab | -0.058 |
| pmp22a | 0.133 | tuba1a | -0.058 |
| crtap | 0.128 | fabp3 | -0.056 |
| cd248b | 0.125 | myt1a | -0.055 |
| cnn2 | 0.125 | myt1b | -0.055 |
| asph | 0.124 | rnasekb | -0.053 |
| fstl1b | 0.120 | si:ch211-137a8.4 | -0.053 |
| lsp1 | 0.119 | cadm3 | -0.051 |
| p3h1 | 0.119 | epb41a | -0.051 |
| pdgfra | 0.119 | fabp7a | -0.050 |
| wls | 0.118 | si:dkey-276j7.1 | -0.050 |
| colec12 | 0.117 | scrt2 | -0.049 |
| twist1a | 0.116 | gapdhs | -0.048 |
| fkbp14 | 0.115 | gng3 | -0.048 |
| fbn2a.1 | 0.115 | insm1a | -0.048 |
| bambia | 0.114 | pvalb1 | -0.046 |
| emp2 | 0.114 | sncb | -0.046 |
| cd151l | 0.112 | CU634008.1 | -0.046 |
| akap12b | 0.111 | CU467822.1 | -0.045 |
| her6 | 0.111 | LOC798783 | -0.045 |
| id3 | 0.111 | atp6v0cb | -0.044 |
| lrrc17 | 0.108 | dlb | -0.044 |
| gpx8 | 0.106 | pou3f1 | -0.044 |
| cthrc1a | 0.105 | tmed4 | -0.044 |
| nid1b | 0.105 | fez1 | -0.043 |
| tgfbi | 0.105 | krtt1c19e | -0.043 |
| XLOC-042222 | 0.105 | pvalb2 | -0.043 |