Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme 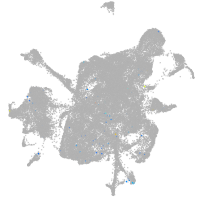 |
hematopoietic / vasculature  |
pronephros 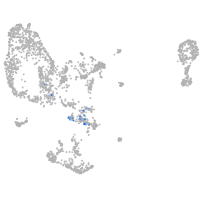 |
neural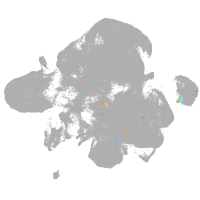 |
spinal cord / glia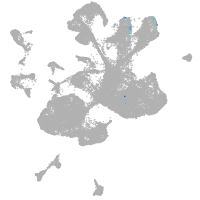 |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ryr1a | 0.291 | gpm6aa | -0.039 |
| mustn1b | 0.287 | hmgb1b | -0.033 |
| myoz2b | 0.285 | ckbb | -0.032 |
| kcnmb2 | 0.275 | mdkb | -0.032 |
| casq2 | 0.272 | chd4a | -0.031 |
| CU633479.2 | 0.269 | h2afva | -0.031 |
| hhatlb | 0.267 | gpm6ab | -0.030 |
| LO018250.1 | 0.265 | si:ch211-288g17.3 | -0.030 |
| rnf207b | 0.263 | si:ch211-137a8.4 | -0.029 |
| myom1b | 0.257 | ddx5 | -0.028 |
| srl | 0.254 | nova2 | -0.028 |
| cavin4a | 0.252 | tuba1c | -0.028 |
| pgam2 | 0.249 | marcksl1a | -0.028 |
| zgc:158296 | 0.247 | setb | -0.027 |
| cavin4b | 0.244 | ywhaqb | -0.027 |
| zgc:86709 | 0.244 | atrx | -0.026 |
| nrap | 0.241 | snrpf | -0.026 |
| si:ch211-28p3.4 | 0.241 | cspg5a | -0.025 |
| cav3 | 0.240 | hdac1 | -0.025 |
| CU681836.1 | 0.240 | pfn2 | -0.025 |
| casq1b | 0.238 | srsf3b | -0.025 |
| klhl43 | 0.236 | stmn1b | -0.025 |
| smyhc2 | 0.235 | cbx3a | -0.024 |
| tnni1c | 0.234 | fabp7a | -0.024 |
| klhl41a | 0.232 | FO082781.1 | -0.024 |
| myot | 0.232 | marcksb | -0.024 |
| tnnc1b | 0.232 | snrpd1 | -0.024 |
| smyhc3 | 0.232 | tubb5 | -0.024 |
| si:ch211-266g18.10 | 0.231 | alyref | -0.023 |
| tmem182a | 0.231 | anp32a | -0.023 |
| si:ch211-157b11.14 | 0.231 | ctsla | -0.023 |
| desma | 0.230 | epb41a | -0.023 |
| mybpc3 | 0.229 | gnb1b | -0.023 |
| myh7bb | 0.229 | nucks1a | -0.023 |
| trdn | 0.229 | seta | -0.023 |




