wu:fc23c09
ZFIN 
Other cell groups


all cells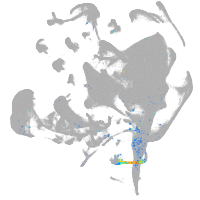 |
blastomeres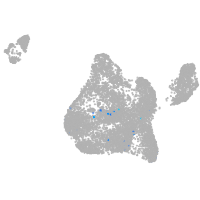 |
PGCs |
endoderm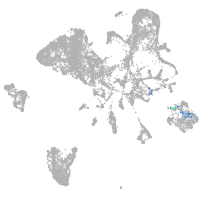 |
axial mesoderm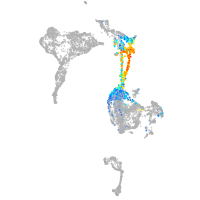 |
muscle 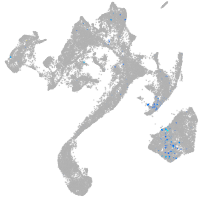 |
mural cells / non-skeletal muscle  |
mesenchyme 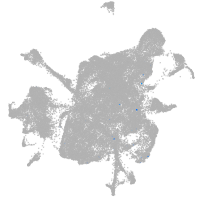 |
hematopoietic / vasculature 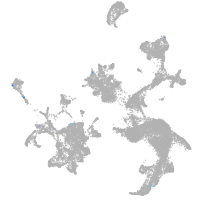 |
pronephros 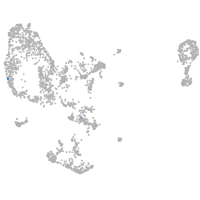 |
neural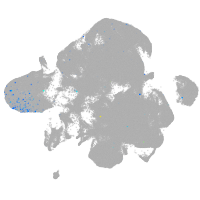 |
spinal cord / glia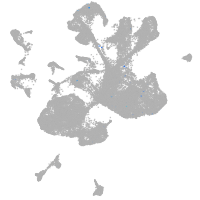 |
eye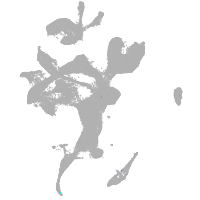 |
taste / olfactory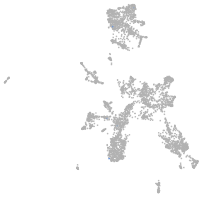 |
otic / lateral line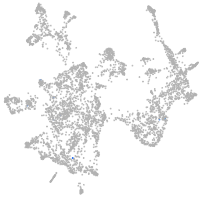 |
epidermis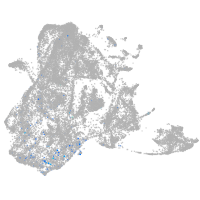 |
periderm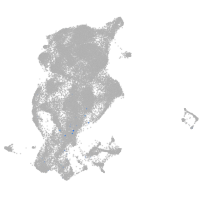 |
fin |
ionocytes / mucous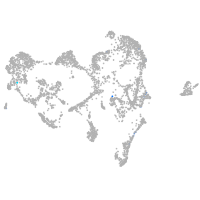 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| paqr4b | 0.361 | prdx2 | -0.046 |
| cmn | 0.342 | tuba1c | -0.045 |
| zmp:0000000760 | 0.331 | ckbb | -0.041 |
| loxl5b | 0.317 | gpm6aa | -0.041 |
| zgc:113232 | 0.308 | pvalb1 | -0.039 |
| col8a1a | 0.302 | rtn1a | -0.039 |
| si:ch73-23l24.1 | 0.289 | pvalb2 | -0.037 |
| col9a1b | 0.281 | stmn1b | -0.037 |
| si:ch211-106h4.6 | 0.270 | marcksl1a | -0.037 |
| chadla | 0.267 | nova2 | -0.036 |
| slc38a8b | 0.267 | fabp3 | -0.035 |
| si:dkeyp-118a3.2 | 0.258 | gpm6ab | -0.035 |
| susd5 | 0.256 | rnasekb | -0.035 |
| tbx3b | 0.253 | elavl3 | -0.034 |
| col5a3a | 0.243 | gng3 | -0.033 |
| sncga | 0.243 | atp6v0cb | -0.032 |
| tbxta | 0.236 | sncb | -0.032 |
| si:ch211-13f8.2 | 0.227 | gapdhs | -0.031 |
| ihhb | 0.217 | hbae3 | -0.031 |
| vgll2b | 0.213 | hbbe1.3 | -0.031 |
| ca8 | 0.204 | mdkb | -0.031 |
| noxo1b | 0.202 | tmsb | -0.031 |
| agtrap | 0.199 | cspg5a | -0.030 |
| ntd5 | 0.196 | sod1 | -0.030 |
| rab38c | 0.194 | vamp2 | -0.030 |
| plod1a | 0.190 | celf2 | -0.029 |
| haus7 | 0.189 | jpt1b | -0.029 |
| egf | 0.188 | tuba1a | -0.029 |
| loxl3b | 0.187 | elavl4 | -0.028 |
| trhra | 0.182 | fez1 | -0.028 |
| golim4b | 0.180 | hbbe1.1 | -0.028 |
| snorc | 0.180 | ndrg2 | -0.028 |
| cnmd | 0.173 | si:dkey-276j7.1 | -0.028 |
| col9a2 | 0.170 | si:dkeyp-75h12.5 | -0.028 |
| shha | 0.169 | dpysl2b | -0.028 |




