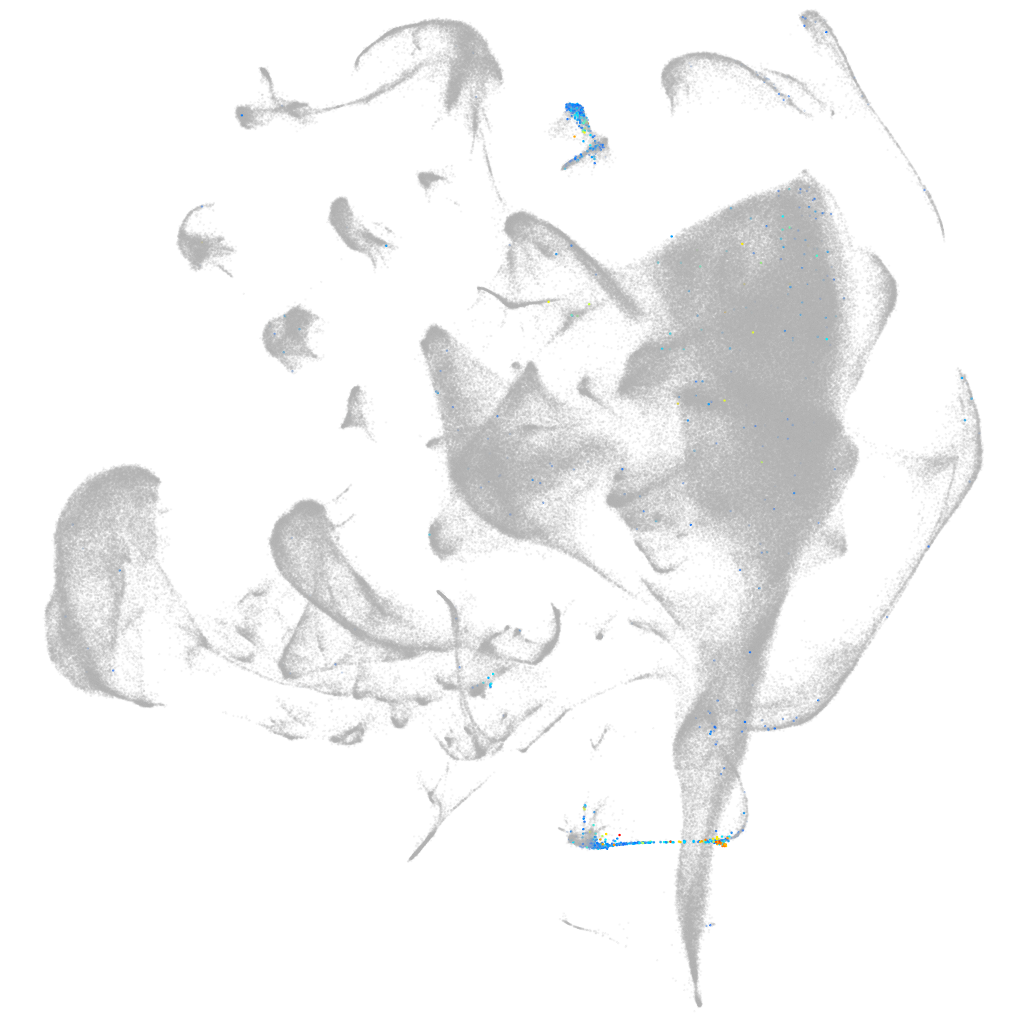
si:dkeyp-118a3.2
ZFIN 
Other cell groups


all cells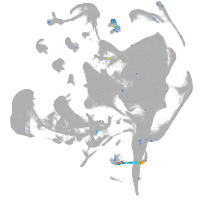 |
blastomeres |
PGCs |
endoderm |
axial mesoderm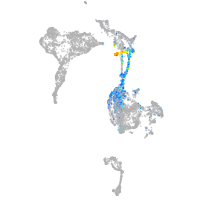 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye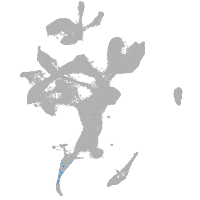 |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| paqr4b | 0.263 | marcksl1a | -0.031 |
| chadla | 0.258 | nova2 | -0.028 |
| wu:fc23c09 | 0.258 | hmgb1b | -0.027 |
| zmp:0000000760 | 0.250 | tuba1c | -0.027 |
| cmn | 0.249 | gpm6aa | -0.026 |
| agtrap | 0.247 | mdkb | -0.026 |
| zgc:113232 | 0.247 | marcksb | -0.025 |
| si:ch211-106h4.6 | 0.238 | hdac1 | -0.024 |
| susd5 | 0.230 | seta | -0.024 |
| col9a1b | 0.225 | si:ch211-288g17.3 | -0.024 |
| si:ch73-23l24.1 | 0.224 | apex1 | -0.023 |
| zgc:91968 | 0.224 | cbx3a | -0.023 |
| si:ch73-389b16.1 | 0.220 | chd4a | -0.023 |
| rab38c | 0.213 | si:dkey-42i9.4 | -0.023 |
| si:ch211-13f8.2 | 0.212 | stmn1a | -0.023 |
| tyrp1a | 0.211 | ddx39aa | -0.022 |
| oca2 | 0.208 | hmgb3a | -0.022 |
| col8a1a | 0.207 | hnrnpub | -0.022 |
| slc24a5 | 0.207 | ilf3b | -0.022 |
| pmela | 0.203 | setb | -0.022 |
| AL935199.1 | 0.201 | elavl3 | -0.021 |
| tyrp1b | 0.198 | epcam | -0.021 |
| dct | 0.195 | h2afva | -0.021 |
| tyr | 0.193 | hmgn3 | -0.021 |
| loxl5b | 0.192 | hnrnpa1b | -0.021 |
| mlpha | 0.191 | tmeff1b | -0.021 |
| mchr2 | 0.190 | tuba1a | -0.021 |
| slc45a2 | 0.182 | mdka | -0.020 |
| slc30a1b | 0.180 | mex3b | -0.020 |
| tspan10 | 0.180 | nono | -0.020 |
| LO018422.1 | 0.176 | si:ch211-137a8.4 | -0.020 |
| sncga | 0.176 | snrpb | -0.020 |
| aadac | 0.172 | sox11b | -0.020 |
| rab38 | 0.169 | stmn1b | -0.020 |
| mitfa | 0.167 | tmpob | -0.020 |