ubiquitin protein ligase E3B
ZFIN 
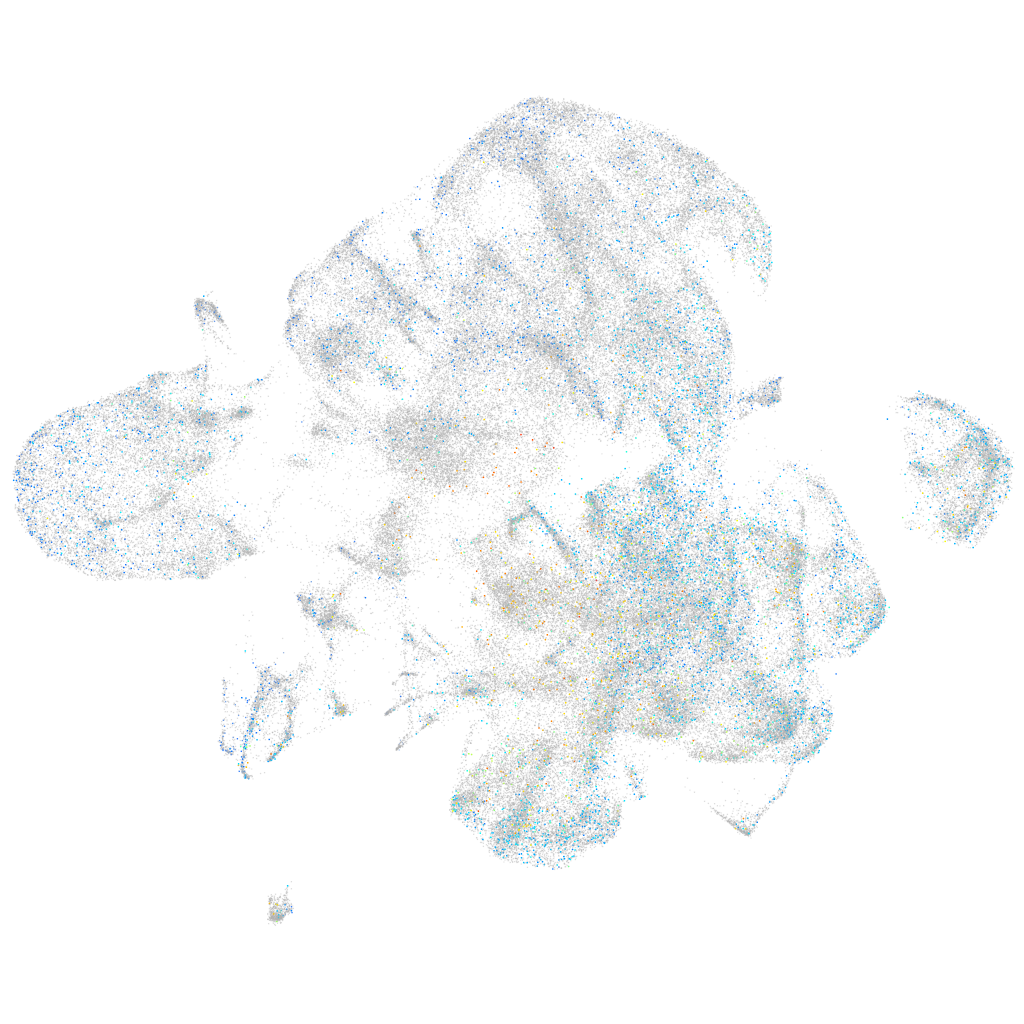
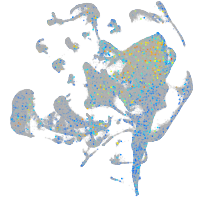
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| elavl3 | 0.085 | hmgb2a | -0.078 |
| rtn1a | 0.078 | pcna | -0.071 |
| stmn1b | 0.078 | stmn1a | -0.069 |
| ywhah | 0.070 | id1 | -0.064 |
| gng3 | 0.070 | selenoh | -0.063 |
| ckbb | 0.069 | dut | -0.062 |
| zc4h2 | 0.069 | tuba8l4 | -0.062 |
| hmgb3a | 0.067 | tuba8l | -0.061 |
| h2afx1 | 0.067 | ranbp1 | -0.061 |
| dpysl3 | 0.067 | nutf2l | -0.060 |
| gpm6aa | 0.066 | si:ch73-281n10.2 | -0.059 |
| myt1b | 0.066 | rrm1 | -0.058 |
| elavl4 | 0.066 | rpa3 | -0.058 |
| tmsb | 0.066 | anp32b | -0.056 |
| tuba1c | 0.065 | cdca7b | -0.055 |
| nova2 | 0.064 | COX7A2 (1 of many) | -0.055 |
| gpm6ab | 0.064 | mdka | -0.055 |
| gng2 | 0.063 | msna | -0.055 |
| tubb5 | 0.063 | fen1 | -0.055 |
| fez1 | 0.062 | snu13b | -0.054 |
| sncb | 0.062 | mcm7 | -0.054 |
| myt1a | 0.062 | lbr | -0.054 |
| vamp2 | 0.062 | chaf1a | -0.054 |
| ptmaa | 0.061 | zgc:56493 | -0.054 |
| jpt1b | 0.061 | npm1a | -0.053 |
| stxbp1a | 0.060 | her2 | -0.053 |
| si:dkey-276j7.1 | 0.060 | mcm5 | -0.053 |
| cbfa2t3 | 0.060 | ccnd1 | -0.053 |
| atp6v1g1 | 0.060 | rrm2 | -0.052 |
| gatad2b | 0.059 | rpa2 | -0.052 |
| atp6v1e1b | 0.058 | ccna2 | -0.052 |
| dbn1 | 0.058 | cks1b | -0.052 |
| celf2 | 0.058 | nasp | -0.052 |
| atp6v0cb | 0.058 | pa2g4b | -0.052 |
| rtn1b | 0.058 | paics | -0.051 |