ubiquitin-conjugating enzyme E2Q family-like 1
ZFIN 
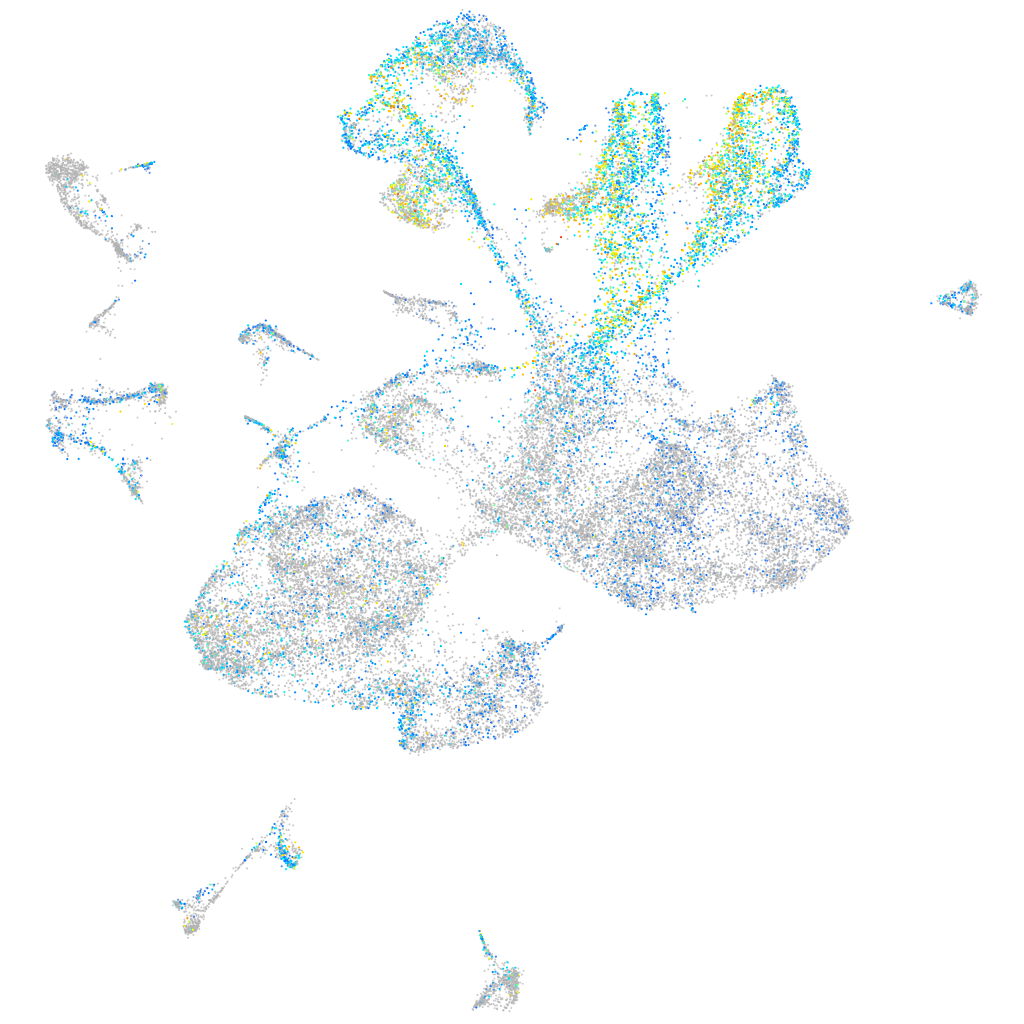
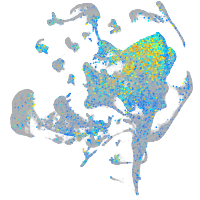
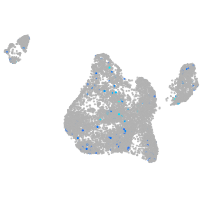
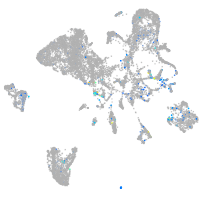
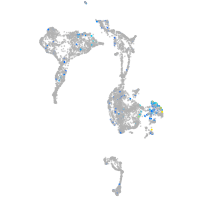
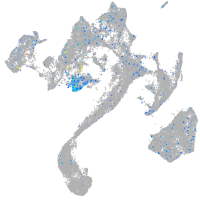
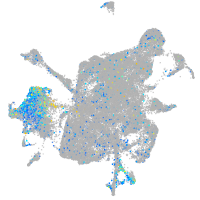
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| stxbp1a | 0.458 | id1 | -0.346 |
| stx1b | 0.457 | XLOC-003690 | -0.325 |
| elavl3 | 0.433 | mdka | -0.318 |
| myt1b | 0.430 | hmgb2a | -0.300 |
| elavl4 | 0.419 | cldn5a | -0.286 |
| cplx2 | 0.419 | sox3 | -0.275 |
| vamp2 | 0.408 | msna | -0.272 |
| snap25a | 0.404 | her15.1 | -0.270 |
| syt2a | 0.403 | notch3 | -0.258 |
| atp6v0cb | 0.401 | vamp3 | -0.258 |
| sncb | 0.401 | XLOC-003689 | -0.247 |
| gng3 | 0.396 | sdc4 | -0.244 |
| ywhah | 0.396 | her12 | -0.241 |
| sncgb | 0.393 | COX7A2 (1 of many) | -0.237 |
| ywhag2 | 0.383 | tuba8l | -0.235 |
| lhx1a | 0.374 | msi1 | -0.234 |
| tmem59l | 0.367 | sox19a | -0.231 |
| atp1a3a | 0.364 | rplp1 | -0.231 |
| cplx2l | 0.359 | pcna | -0.228 |
| nsg2 | 0.358 | si:dkey-151g10.6 | -0.227 |
| atp1b2a | 0.358 | si:ch73-281n10.2 | -0.225 |
| gng2 | 0.357 | zgc:56493 | -0.223 |
| stmn2a | 0.352 | dut | -0.223 |
| eno2 | 0.351 | selenoh | -0.222 |
| rtn1b | 0.351 | btg2 | -0.219 |
| rbfox3a | 0.351 | tuba8l4 | -0.218 |
| rtn1a | 0.349 | cd82a | -0.218 |
| rbfox1 | 0.348 | her2 | -0.217 |
| nova1 | 0.342 | sparc | -0.216 |
| nrxn1a | 0.341 | fosab | -0.216 |
| gjd1a | 0.341 | stmn1a | -0.216 |
| jagn1a | 0.340 | ranbp1 | -0.215 |
| csdc2a | 0.340 | her4.1 | -0.210 |
| stmn1b | 0.339 | gas1b | -0.208 |
| kcnc3a | 0.339 | ccng1 | -0.207 |