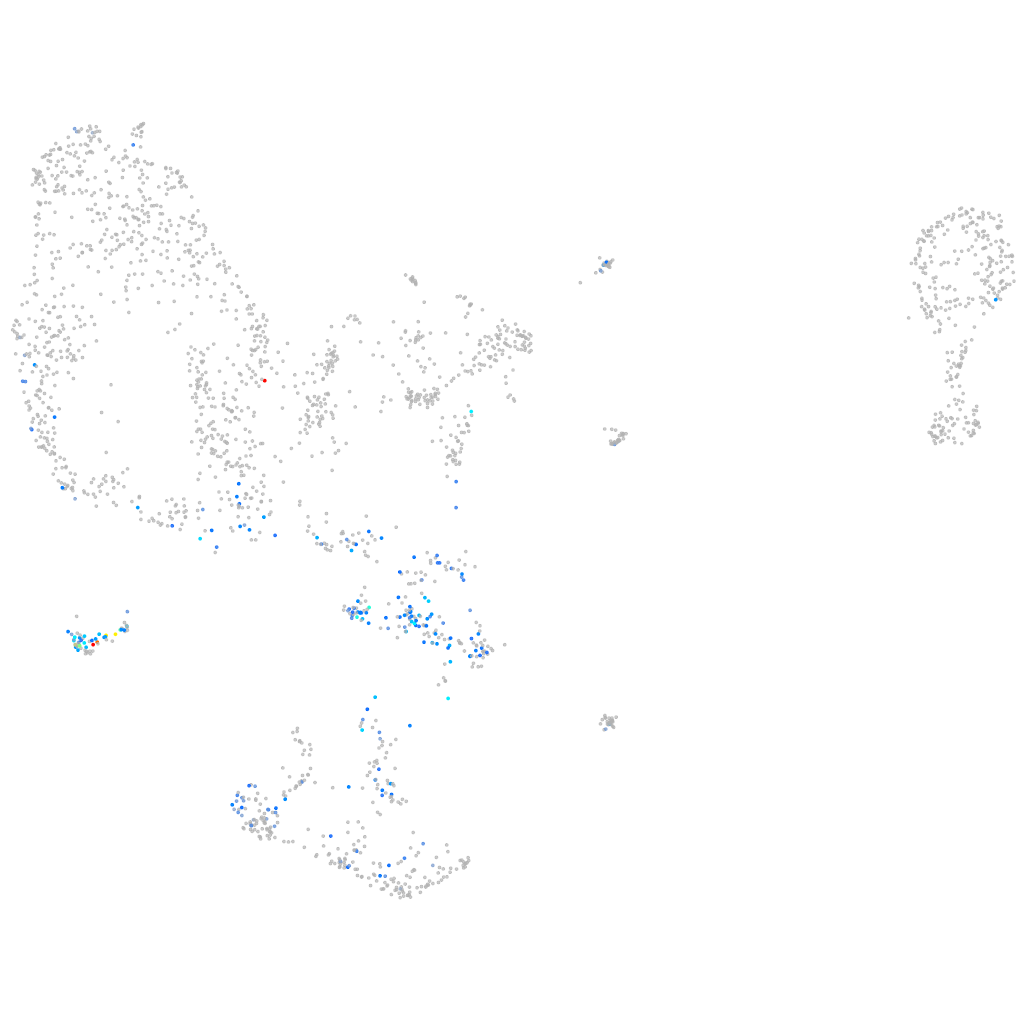
"testis specific, 10"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| spata18 | 0.462 | gapdh | -0.175 |
| rsph9 | 0.417 | rplp2 | -0.169 |
| ccdc151 | 0.403 | gcshb | -0.165 |
| LOC100329490 | 0.397 | dab2 | -0.164 |
| dnaaf3 | 0.394 | grhprb | -0.162 |
| zgc:55461 | 0.390 | aldob | -0.159 |
| cetn4 | 0.388 | hao1 | -0.156 |
| si:ch1073-416d2.4 | 0.387 | dpys | -0.156 |
| si:ch211-163l21.7 | 0.386 | cubn | -0.155 |
| cdc25d | 0.384 | pnp6 | -0.155 |
| irak1bp1 | 0.384 | fbp1b | -0.154 |
| spaca9 | 0.379 | si:dkey-33i11.9 | -0.153 |
| cfap45 | 0.379 | upb1 | -0.152 |
| cfap20 | 0.378 | nit2 | -0.152 |
| cluap1 | 0.377 | ahcy | -0.151 |
| tekt2 | 0.373 | gamt | -0.151 |
| iqcd | 0.373 | alpl | -0.150 |
| ribc2 | 0.368 | zgc:136493 | -0.147 |
| spata4 | 0.368 | rpl3 | -0.145 |
| cfap58 | 0.361 | pdzk1ip1 | -0.143 |
| dcdc2b | 0.361 | epdl2 | -0.141 |
| erich3 | 0.360 | gstt1a | -0.141 |
| ccdc173 | 0.358 | slc26a1 | -0.141 |
| tex9 | 0.358 | dpydb | -0.140 |
| dnali1 | 0.355 | ctsz | -0.140 |
| mycbp | 0.353 | phyhd1 | -0.139 |
| iqcg | 0.352 | cx32.3 | -0.138 |
| catip | 0.352 | rps18 | -0.138 |
| ttc25 | 0.351 | si:dkey-28n18.9 | -0.137 |
| ropn1l | 0.351 | gpt2l | -0.137 |
| dnal1 | 0.349 | nipsnap3a | -0.136 |
| foxj1a | 0.348 | lrp2a | -0.136 |
| odf3l2b | 0.348 | ASS1 | -0.135 |
| tppp3 | 0.346 | sord | -0.135 |
| dzip1l | 0.344 | aldh7a1 | -0.135 |