"testis specific, 10"
ZFIN 
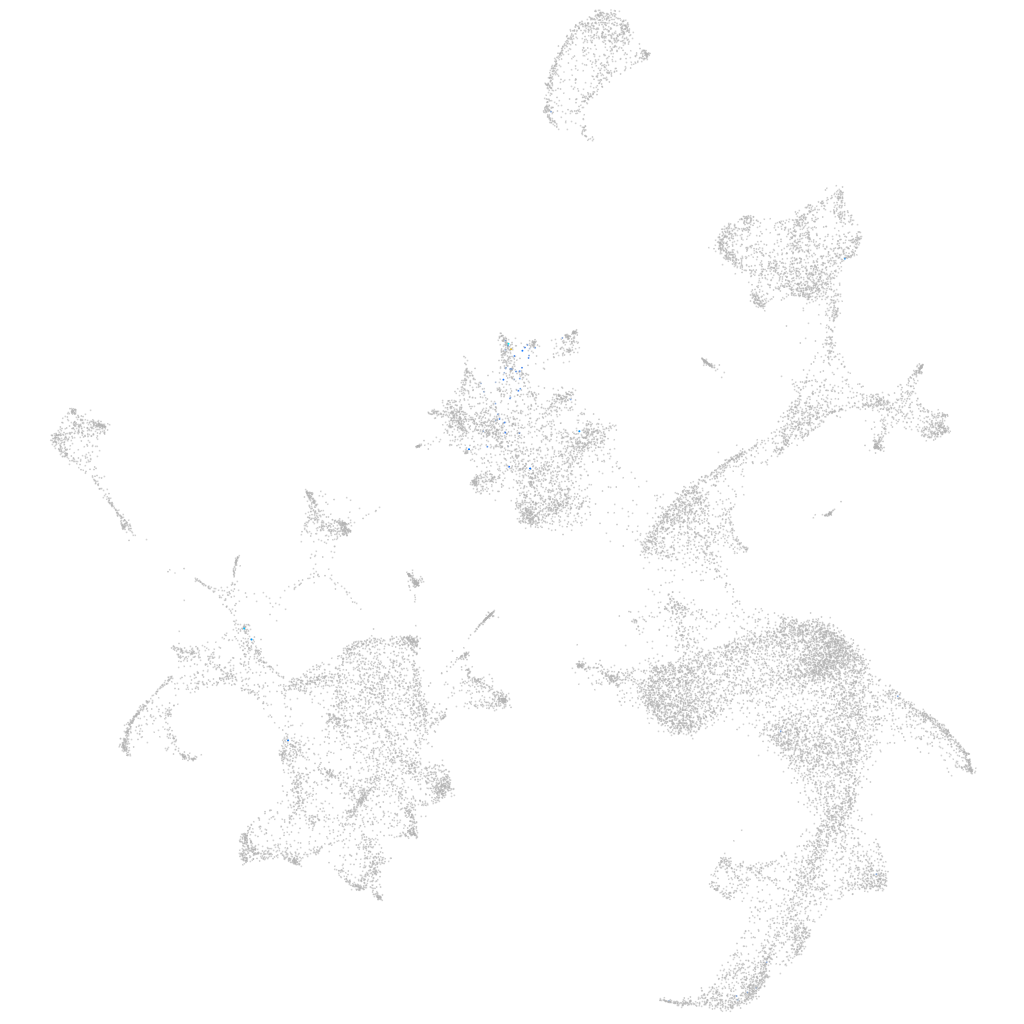
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm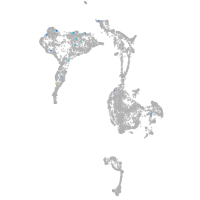 |
muscle 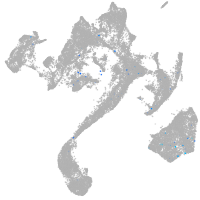 |
mural cells / non-skeletal muscle  |
mesenchyme 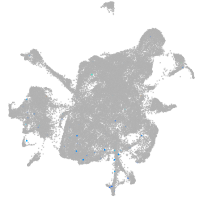 |
hematopoietic / vasculature  |
pronephros 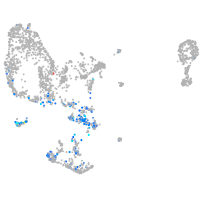 |
neural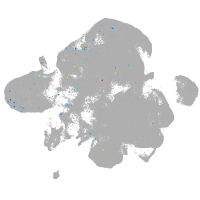 |
spinal cord / glia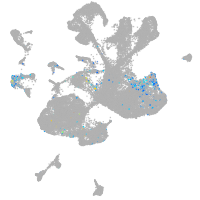 |
eye |
taste / olfactory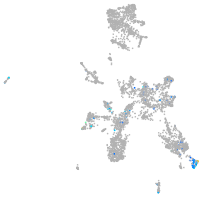 |
otic / lateral line |
epidermis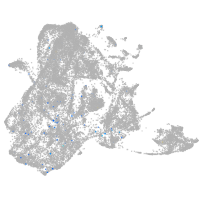 |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cnga3b | 0.300 | arhgdia | -0.017 |
| si:ch73-6k14.2 | 0.280 | pvalb1 | -0.016 |
| prph2l | 0.258 | cdh5 | -0.016 |
| XLOC-005660 | 0.258 | pfn1 | -0.016 |
| rcvrna | 0.253 | fli1a | -0.016 |
| gngt2a | 0.252 | tie1 | -0.015 |
| reep6 | 0.250 | tspan14 | -0.014 |
| si:dkey-72l14.3 | 0.242 | b3gnt3.4 | -0.014 |
| tmem244 | 0.241 | mfng | -0.014 |
| arl13a | 0.230 | ehd4 | -0.014 |
| TULP1 | 0.227 | clec14a | -0.014 |
| tulp1a | 0.227 | pecam1 | -0.014 |
| rgs9a | 0.226 | podxl | -0.013 |
| si:ch73-28h20.1 | 0.225 | yrk | -0.013 |
| mpp4b | 0.225 | pvalb2 | -0.013 |
| zmp:0000000545 | 0.223 | crip1 | -0.013 |
| rx2 | 0.222 | arhgdig | -0.013 |
| arl3l1 | 0.219 | si:dkey-27i16.2 | -0.013 |
| si:dkey-283b15.2 | 0.219 | abracl | -0.013 |
| si:dkey-220f10.4 | 0.213 | kdr | -0.013 |
| rcvrn2 | 0.213 | rac2 | -0.013 |
| ppa1a | 0.212 | snx9b | -0.012 |
| atp2b1b | 0.210 | XLOC-014079 | -0.012 |
| elovl4b | 0.209 | mylz3 | -0.012 |
| guk1b | 0.207 | pycard | -0.012 |
| ldhbb | 0.206 | crema | -0.012 |
| tmx3a | 0.206 | myhz1.1 | -0.012 |
| zgc:109965 | 0.204 | wasa | -0.012 |
| LOC103909376 | 0.201 | ucp2 | -0.012 |
| ntng2a | 0.201 | XLOC-039757 | -0.012 |
| tulp1b | 0.201 | rab8b | -0.012 |
| foxq2 | 0.201 | coro1ca | -0.012 |
| si:dkey-30k22.7 | 0.199 | myct1a | -0.012 |
| ugt2a6 | 0.199 | mcamb | -0.012 |
| CABZ01015105.1 | 0.197 | gpr182 | -0.012 |