trichorhinophalangeal syndrome I
ZFIN 
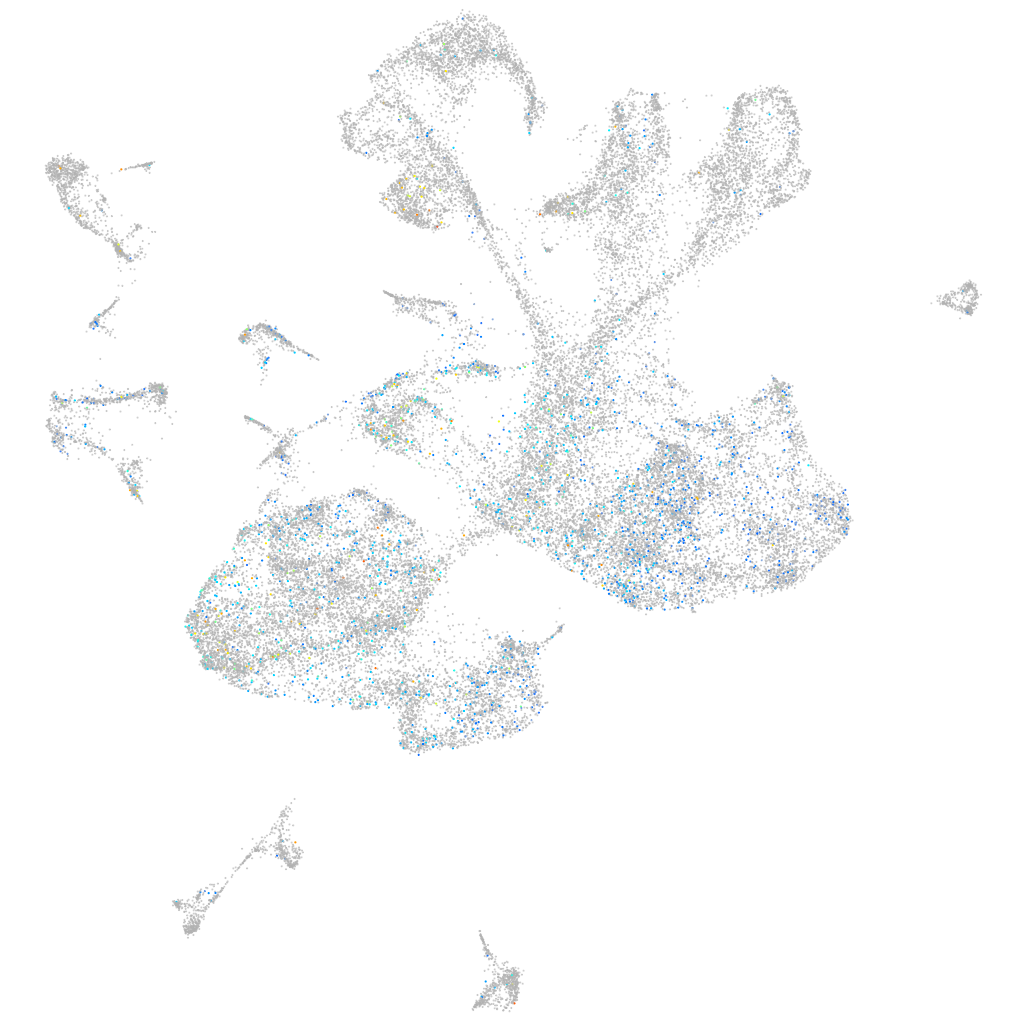
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-251b21.1 | 0.087 | elavl3 | -0.067 |
| atp1a1b | 0.085 | stxbp1a | -0.066 |
| prss35 | 0.083 | gng3 | -0.064 |
| id1 | 0.082 | vamp2 | -0.062 |
| atp1b4 | 0.082 | jagn1a | -0.059 |
| slc6a9 | 0.080 | sncb | -0.059 |
| endouc | 0.078 | nsg2 | -0.058 |
| zgc:165461 | 0.075 | atp6v0cb | -0.058 |
| XLOC-003690 | 0.074 | stx1b | -0.058 |
| sox19a | 0.074 | gng2 | -0.057 |
| ptprfb | 0.074 | stmn1b | -0.057 |
| slc6a11b | 0.073 | ptmaa | -0.055 |
| ptprz1b | 0.073 | myt1b | -0.055 |
| notch3 | 0.073 | snap25a | -0.054 |
| msi1 | 0.073 | ywhag2 | -0.054 |
| sox3 | 0.072 | tubb5 | -0.054 |
| fabp7a | 0.071 | cplx2l | -0.053 |
| wnt7aa | 0.069 | tmem59l | -0.053 |
| mdkb | 0.069 | calm1a | -0.053 |
| fam181b | 0.069 | si:dkey-276j7.1 | -0.052 |
| sparc | 0.069 | stmn2a | -0.052 |
| gfap | 0.068 | elavl4 | -0.052 |
| mdka | 0.067 | rtn1a | -0.051 |
| gpm6bb | 0.067 | rbfox1 | -0.051 |
| pax3a | 0.066 | cnrip1a | -0.050 |
| hepacama | 0.065 | onecut1 | -0.050 |
| sox2 | 0.065 | cplx2 | -0.050 |
| col4a5 | 0.065 | ywhah | -0.049 |
| si:ch1073-303k11.2 | 0.065 | zgc:65894 | -0.049 |
| boc | 0.065 | fscn1a | -0.049 |
| sema4e | 0.065 | thsd7aa | -0.048 |
| gas1b | 0.065 | pcsk1nl | -0.048 |
| tmem47 | 0.064 | atcaya | -0.047 |
| angptl4 | 0.064 | aplp1 | -0.047 |
| fgfr2 | 0.063 | gpm6ab | -0.047 |