tRNA selenocysteine 1 associated protein 1a
ZFIN 
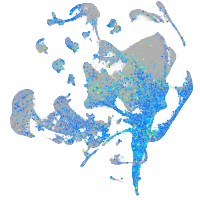
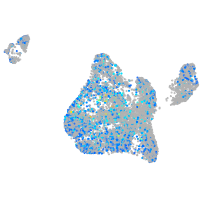
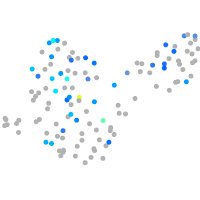
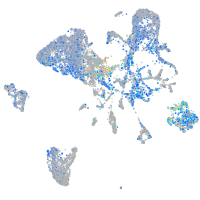
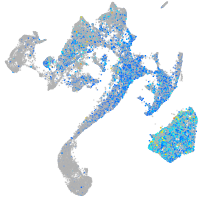
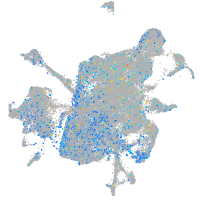
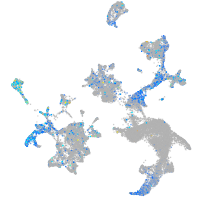
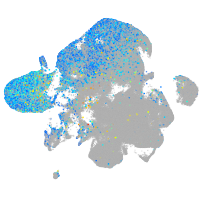
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| npm1a | 0.298 | col2a1a | -0.261 |
| nop58 | 0.292 | rpl37 | -0.259 |
| ncl | 0.289 | col11a1a | -0.253 |
| nop56 | 0.285 | col9a1b | -0.253 |
| dkc1 | 0.281 | sparc | -0.252 |
| nop2 | 0.278 | col9a3 | -0.250 |
| fbl | 0.277 | zgc:114188 | -0.250 |
| snu13b | 0.274 | rps10 | -0.250 |
| ebna1bp2 | 0.265 | col9a2 | -0.245 |
| gnl3 | 0.265 | zmp:0000000760 | -0.245 |
| rsl1d1 | 0.261 | cnmd | -0.242 |
| hmga1a | 0.260 | si:ch73-23l24.1 | -0.240 |
| lyar | 0.260 | cmn | -0.238 |
| ilf3b | 0.260 | col8a1a | -0.234 |
| hspb1 | 0.259 | zgc:113232 | -0.232 |
| pes | 0.256 | rps17 | -0.225 |
| seta | 0.255 | col11a2 | -0.218 |
| syncrip | 0.253 | epyc | -0.205 |
| eif4bb | 0.252 | col2a1b | -0.202 |
| ppig | 0.252 | zgc:162730 | -0.201 |
| hnrnpabb | 0.251 | selenom | -0.196 |
| hnrnpa1a | 0.250 | ctgfa | -0.195 |
| si:ch211-217k17.7 | 0.249 | agtrap | -0.195 |
| cirbpa | 0.249 | ucmab | -0.194 |
| anp32e | 0.246 | loxl1 | -0.194 |
| NC-002333.4 | 0.246 | haus7 | -0.191 |
| setb | 0.246 | slc3a2a | -0.190 |
| hnrnpa1b | 0.246 | tgfb3 | -0.189 |
| ddx18 | 0.246 | krt8 | -0.188 |
| snrnp70 | 0.244 | zgc:158463 | -0.183 |
| khdrbs1a | 0.241 | CABZ01072614.1 | -0.182 |
| snrpb | 0.240 | susd5 | -0.181 |
| bms1 | 0.240 | matn3a | -0.179 |
| hnrnpub | 0.239 | snorc | -0.178 |
| nhp2 | 0.238 | CR383676.1 | -0.178 |