thyroid hormone receptor interactor 13
ZFIN 
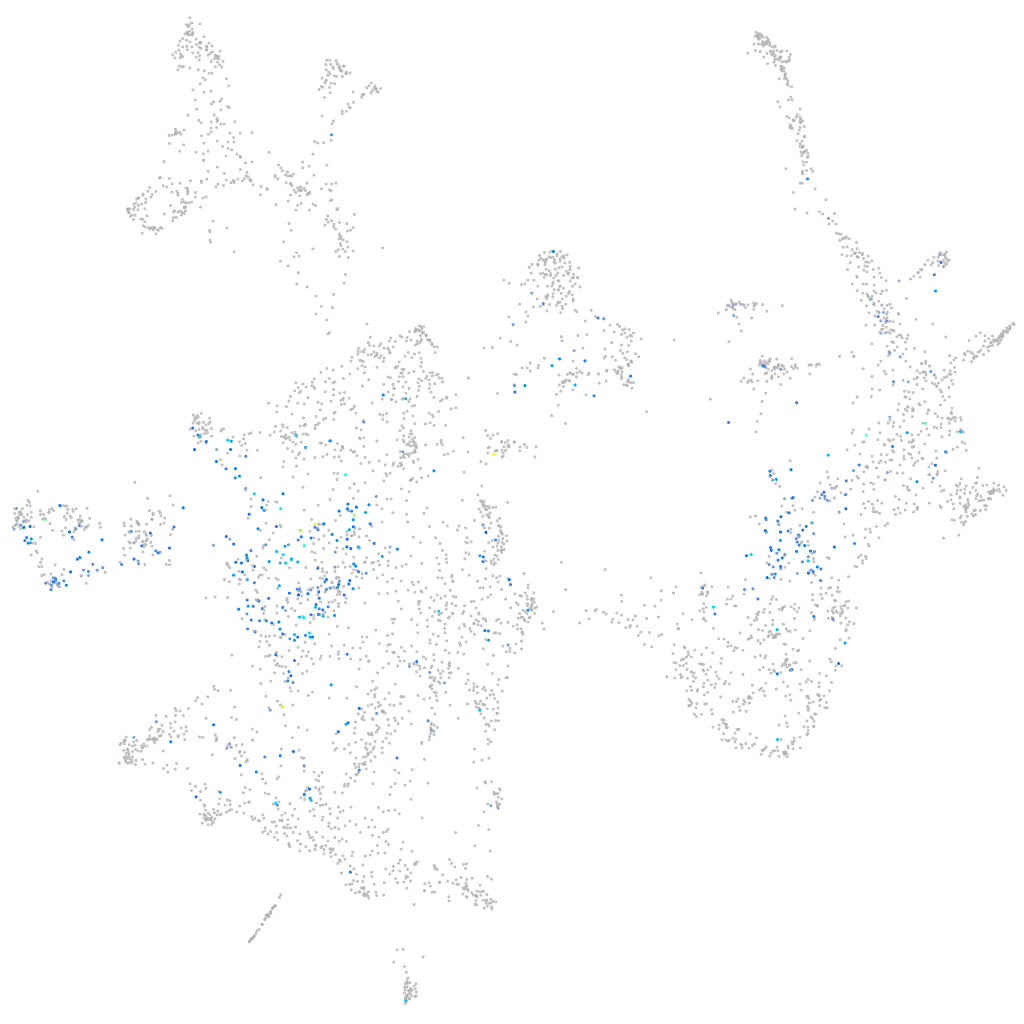
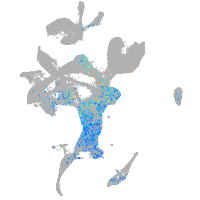
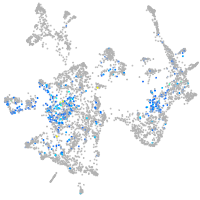
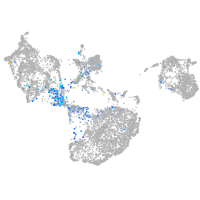
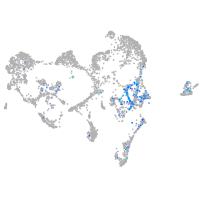
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cks1b | 0.485 | tob1b | -0.138 |
| zgc:110540 | 0.464 | COX3 | -0.137 |
| ccna2 | 0.459 | h1f0 | -0.127 |
| rrm2 | 0.455 | tmem59 | -0.121 |
| rrm1 | 0.448 | atp2b1a | -0.120 |
| dhfr | 0.448 | calm1a | -0.120 |
| cdca5 | 0.442 | nupr1a | -0.119 |
| dut | 0.440 | mt-co2 | -0.118 |
| pcna | 0.433 | b2ml | -0.117 |
| mibp | 0.425 | CR383676.1 | -0.115 |
| aurkb | 0.424 | otofb | -0.115 |
| ncapg | 0.420 | pvalb8 | -0.112 |
| cdk1 | 0.415 | calml4a | -0.111 |
| chaf1a | 0.414 | kif1aa | -0.110 |
| si:dkey-6i22.5 | 0.414 | nptna | -0.109 |
| mad2l1 | 0.413 | gpx2 | -0.109 |
| rpa2 | 0.412 | osbpl1a | -0.109 |
| stmn1a | 0.409 | cd164l2 | -0.108 |
| mki67 | 0.401 | gapdhs | -0.107 |
| fbxo5 | 0.400 | atp1a3b | -0.106 |
| ncapd2 | 0.399 | CR925719.1 | -0.106 |
| dek | 0.390 | rnasekb | -0.105 |
| CABZ01005379.1 | 0.390 | lrrc73 | -0.105 |
| esco2 | 0.381 | ip6k2a | -0.105 |
| tubb2b | 0.379 | pcsk5a | -0.104 |
| slbp | 0.375 | si:ch73-15n15.3 | -0.104 |
| zgc:165555.3 | 0.372 | emb | -0.103 |
| zgc:110216 | 0.371 | tspan13b | -0.101 |
| arhgef39 | 0.368 | dnajc5b | -0.101 |
| shmt1 | 0.367 | pvalb2 | -0.101 |
| mis12 | 0.367 | atp6v1e1b | -0.101 |
| prim2 | 0.366 | prr15la | -0.100 |
| lbr | 0.365 | gipc3 | -0.100 |
| nasp | 0.363 | capgb | -0.100 |
| smc2 | 0.360 | ckbb | -0.099 |