thyroid hormone receptor interactor 13
ZFIN 
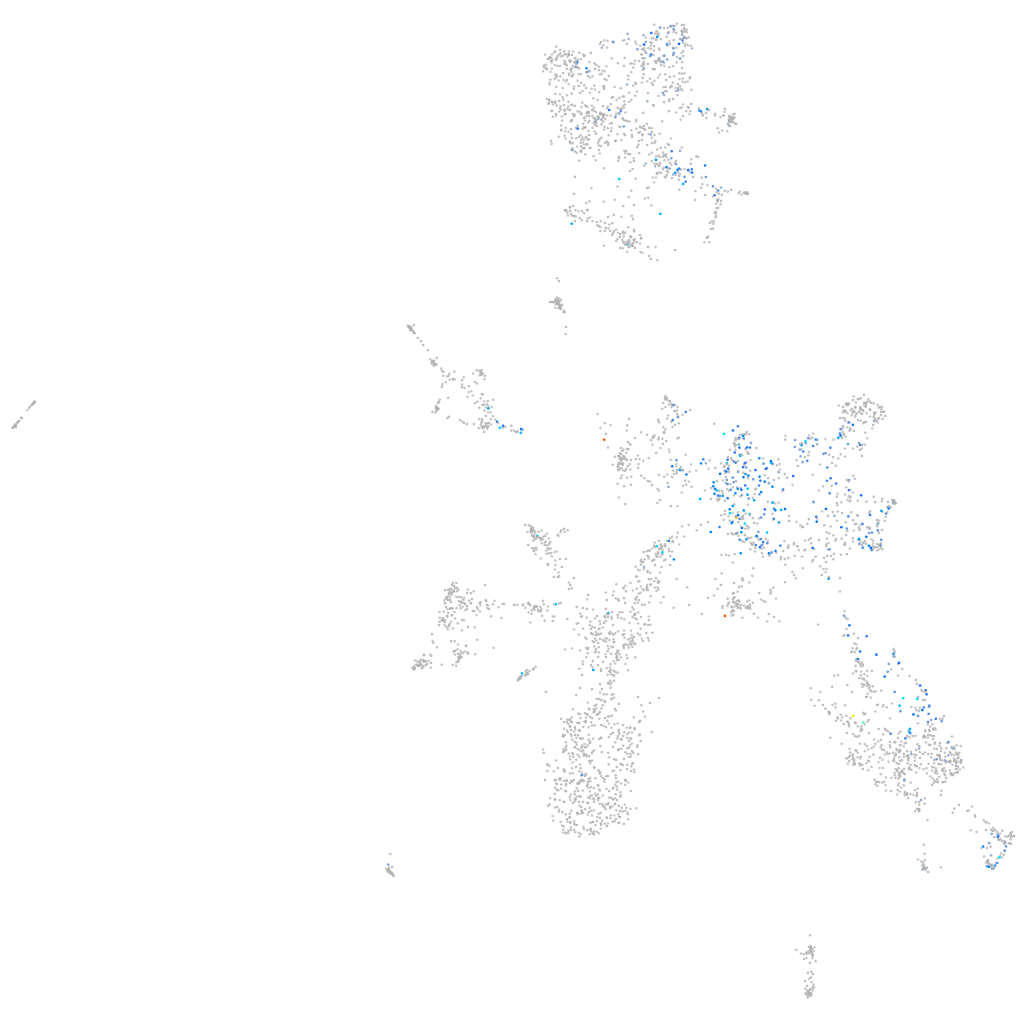
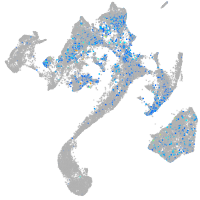
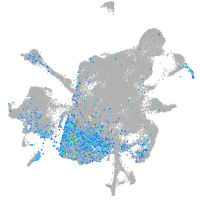
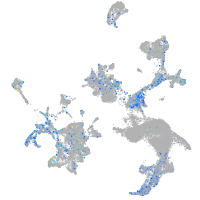
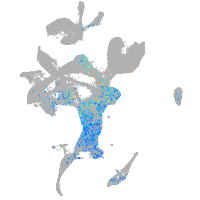
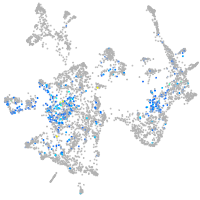
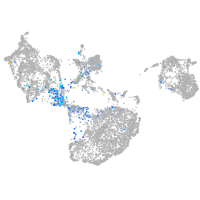
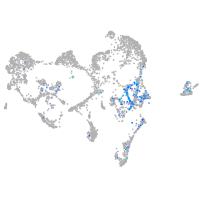
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cks1b | 0.391 | txn | -0.189 |
| zgc:110540 | 0.387 | CR383676.1 | -0.181 |
| stmn1a | 0.385 | anxa5b | -0.169 |
| rrm1 | 0.373 | tmem59 | -0.168 |
| dut | 0.372 | gstp1 | -0.164 |
| rrm2 | 0.372 | calm1a | -0.163 |
| ccna2 | 0.368 | rnasekb | -0.163 |
| esco2 | 0.351 | chga | -0.158 |
| aurkb | 0.350 | atp6v0cb | -0.157 |
| mki67 | 0.348 | vamp2 | -0.155 |
| si:ch211-156b7.4 | 0.330 | gapdhs | -0.152 |
| cdk1 | 0.330 | itm2ba | -0.151 |
| lbr | 0.329 | cldnh | -0.151 |
| mibp | 0.325 | rtn1a | -0.150 |
| cdca5 | 0.325 | camk2n1a | -0.149 |
| pcna | 0.325 | sncb | -0.146 |
| mis12 | 0.323 | bik | -0.145 |
| mad2l1 | 0.322 | syt1a | -0.145 |
| rpa2 | 0.322 | atp6v1e1b | -0.145 |
| si:dkey-6i22.5 | 0.319 | selenow2b | -0.144 |
| zgc:110216 | 0.318 | gnb1a | -0.144 |
| tyms | 0.316 | pvalb5 | -0.144 |
| chaf1a | 0.315 | scg2b | -0.143 |
| ncapg | 0.315 | gabarapa | -0.143 |
| smc2 | 0.314 | si:dkeyp-75h12.5 | -0.139 |
| fbxo5 | 0.314 | atp6v1g1 | -0.138 |
| pbk | 0.313 | tspan2a | -0.137 |
| ndc80 | 0.313 | uchl1 | -0.137 |
| smc4 | 0.311 | nfe2l2a | -0.137 |
| CABZ01005379.1 | 0.307 | zgc:65894 | -0.136 |
| nasp | 0.305 | atp1b1a | -0.136 |
| cdca8 | 0.304 | scg2a | -0.135 |
| dek | 0.301 | COX3 | -0.135 |
| ncaph | 0.300 | zgc:165573 | -0.135 |
| haus4 | 0.300 | gng3 | -0.135 |