thyroid hormone receptor interactor 10a
ZFIN 
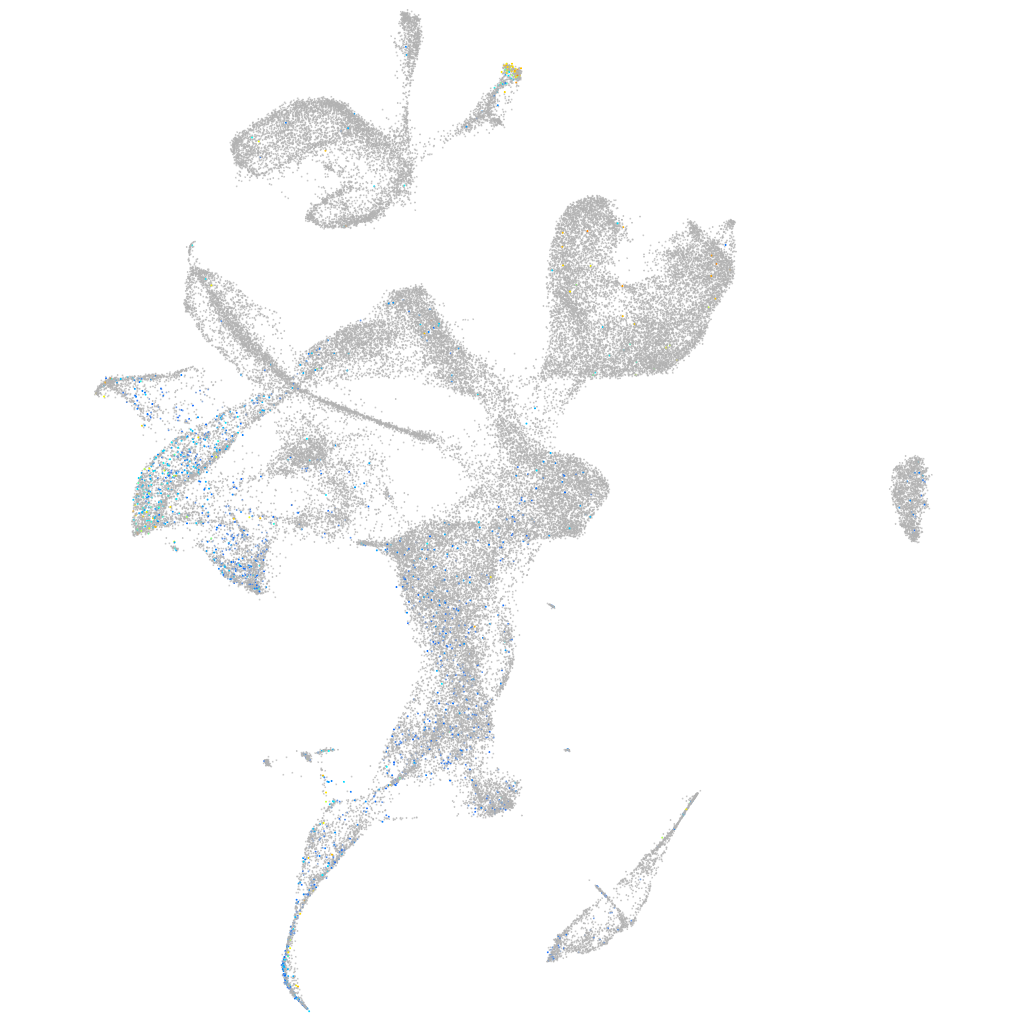
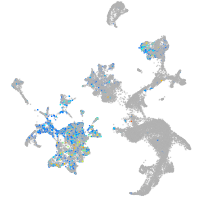
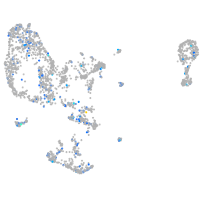
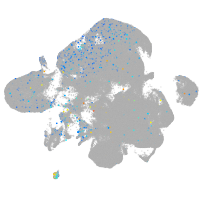
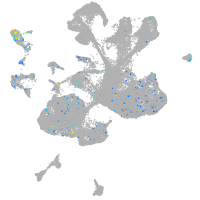
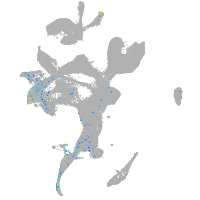
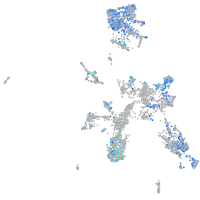
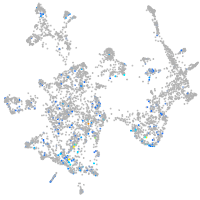
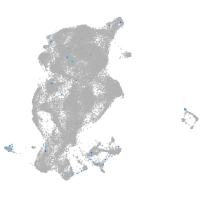
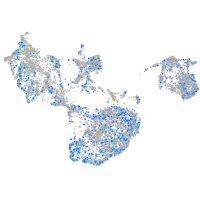
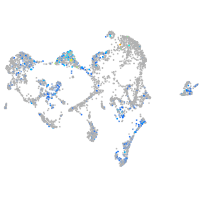
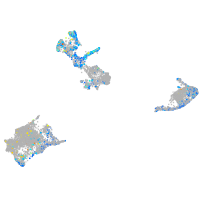
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| BX897741.1 | 0.157 | ptmab | -0.103 |
| rbp4l | 0.154 | marcksl1b | -0.102 |
| gnat2 | 0.151 | cirbpb | -0.097 |
| prph2a | 0.149 | hmgn2 | -0.096 |
| rcvrn3 | 0.147 | hnrnpaba | -0.090 |
| gnb3b | 0.147 | h2afvb | -0.081 |
| zgc:73359 | 0.145 | si:ch211-222l21.1 | -0.077 |
| slc4a5b | 0.144 | hmgb1b | -0.077 |
| si:ch211-81a5.8 | 0.143 | h3f3d | -0.077 |
| pdcb | 0.141 | hmga1a | -0.075 |
| rbp3 | 0.141 | hmgb1a | -0.075 |
| pde6c | 0.140 | si:ch211-288g17.3 | -0.074 |
| grk1b | 0.139 | gpm6aa | -0.073 |
| slc25a3a | 0.139 | si:ch73-1a9.3 | -0.072 |
| ompa | 0.138 | khdrbs1a | -0.071 |
| grk7a | 0.137 | marcksb | -0.071 |
| gngt2b | 0.137 | hnrnpa0b | -0.070 |
| si:ch211-207l14.1 | 0.137 | hnrnpa0l | -0.070 |
| cox4i2 | 0.137 | ptmaa | -0.067 |
| arr3a | 0.137 | nova2 | -0.067 |
| rprmb | 0.135 | tuba1a | -0.067 |
| guca1c | 0.134 | chd4a | -0.066 |
| si:ch1073-303d10.1 | 0.134 | marcksl1a | -0.066 |
| rgs9a | 0.133 | hsp90ab1 | -0.064 |
| si:dkey-17e16.15 | 0.132 | hmgb2b | -0.064 |
| pde6ha | 0.132 | hmgb3a | -0.064 |
| guk1b | 0.131 | smarce1 | -0.063 |
| prph2b | 0.131 | h2afva | -0.062 |
| inaa | 0.128 | cfl1 | -0.060 |
| si:ch1073-469d17.2 | 0.128 | hnrnpabb | -0.058 |
| pde6h | 0.125 | si:dkey-276j7.1 | -0.058 |
| ppdpfa | 0.125 | zc4h2 | -0.058 |
| plekhd1 | 0.125 | tubb2b | -0.057 |
| eno1b | 0.124 | sumo3a | -0.056 |
| rcvrn2 | 0.124 | hdac1 | -0.056 |