trafficking protein particle complex 6b
ZFIN 
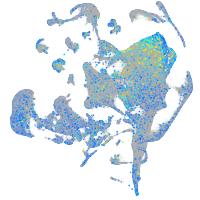
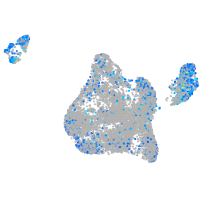
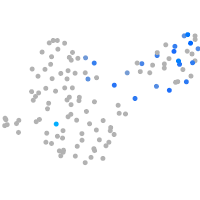
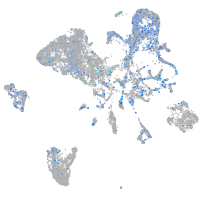
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| stmn2a | 0.132 | hmgb2a | -0.092 |
| rtn1b | 0.128 | pcna | -0.055 |
| zgc:65894 | 0.127 | lig1 | -0.053 |
| rbpms2a | 0.124 | chaf1a | -0.053 |
| mllt11 | 0.122 | rrm1 | -0.050 |
| elavl3 | 0.122 | mki67 | -0.049 |
| islr2 | 0.120 | ccnd1 | -0.049 |
| ywhag2 | 0.119 | stmn1a | -0.048 |
| gng3 | 0.118 | her15.1 | -0.048 |
| rbpms2b | 0.117 | dek | -0.048 |
| eno2 | 0.116 | CABZ01005379.1 | -0.046 |
| zgc:153426 | 0.115 | banf1 | -0.045 |
| stxbp1a | 0.115 | mcm7 | -0.045 |
| sncb | 0.114 | mibp | -0.044 |
| gap43 | 0.113 | fen1 | -0.044 |
| ppp1r14ba | 0.113 | nasp | -0.044 |
| cplx2l | 0.110 | rrm2 | -0.044 |
| nrn1a | 0.110 | lbr | -0.043 |
| id4 | 0.109 | mcm6 | -0.043 |
| inab | 0.108 | zgc:110425 | -0.043 |
| isl2b | 0.108 | nutf2l | -0.043 |
| stmn1b | 0.107 | hmgb2b | -0.042 |
| gnao1a | 0.106 | smc4 | -0.042 |
| snap25a | 0.105 | rpa3 | -0.042 |
| stmn2b | 0.102 | dut | -0.042 |
| elavl4 | 0.101 | ccna2 | -0.042 |
| si:dkeyp-75h12.5 | 0.100 | dlgap5 | -0.042 |
| tubb5 | 0.100 | hmga1a | -0.042 |
| mab21l1 | 0.100 | anp32b | -0.042 |
| stx1b | 0.099 | zgc:110216 | -0.041 |
| tuba2 | 0.099 | neurod4 | -0.041 |
| si:dkey-280e21.3 | 0.098 | si:ch73-281n10.2 | -0.041 |
| xpr1a | 0.098 | mcm3 | -0.041 |
| carmil2 | 0.098 | shisa2a | -0.041 |
| zfhx3 | 0.098 | hes2.2 | -0.041 |