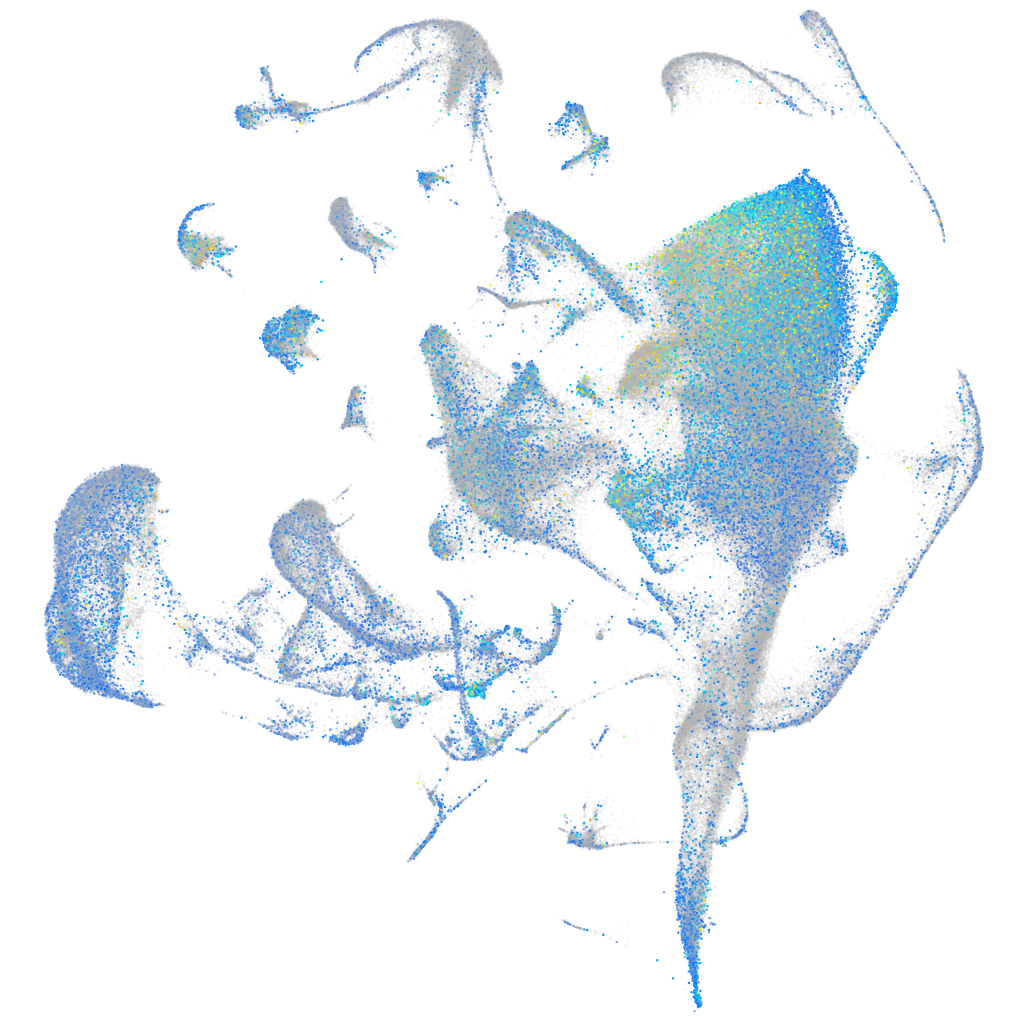trafficking protein particle complex 6b
ZFIN 
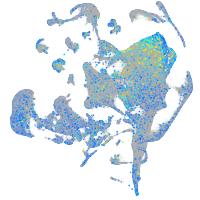
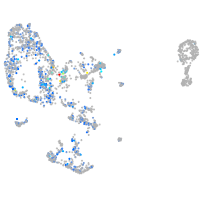
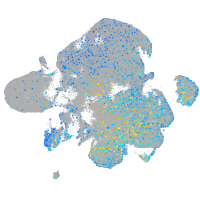
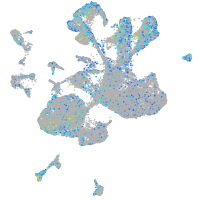
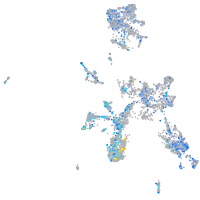
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gng3 | 0.144 | hspb1 | -0.073 |
| mllt11 | 0.137 | apoc1 | -0.062 |
| sncb | 0.137 | dkc1 | -0.061 |
| stmn2a | 0.137 | nop2 | -0.061 |
| zgc:65894 | 0.137 | si:ch211-152c2.3 | -0.061 |
| ywhag2 | 0.133 | fbl | -0.060 |
| atp6v1e1b | 0.128 | nop58 | -0.060 |
| atp6v1g1 | 0.128 | npm1a | -0.060 |
| stxbp1a | 0.128 | lig1 | -0.059 |
| calm2a | 0.127 | rsl1d1 | -0.059 |
| ywhah | 0.126 | mki67 | -0.058 |
| gap43 | 0.125 | zmp:0000000624 | -0.058 |
| calm1a | 0.125 | nop56 | -0.056 |
| stmn1b | 0.124 | si:dkey-66i24.9 | -0.056 |
| gnb1a | 0.123 | zgc:110425 | -0.056 |
| rtn1b | 0.122 | sp5l | -0.055 |
| snap25a | 0.122 | anp32b | -0.054 |
| gnao1a | 0.121 | chaf1a | -0.054 |
| vamp2 | 0.121 | gnl3 | -0.054 |
| gng2 | 0.119 | apoeb | -0.053 |
| tuba1c | 0.119 | ccnd1 | -0.053 |
| fez1 | 0.118 | cx43.4 | -0.052 |
| stx1b | 0.118 | ddx18 | -0.052 |
| elavl4 | 0.117 | pcna | -0.052 |
| gdi1 | 0.117 | stm | -0.052 |
| id4 | 0.117 | id1 | -0.051 |
| si:dkeyp-75h12.5 | 0.117 | mcm6 | -0.051 |
| tmsb2 | 0.116 | si:dkey-239h2.3 | -0.051 |
| cnrip1a | 0.114 | bms1 | -0.050 |
| csdc2a | 0.114 | cdx4 | -0.050 |
| rnasekb | 0.114 | dnmt3bb.2 | -0.050 |
| rtn1a | 0.114 | mcm3 | -0.050 |
| zgc:153426 | 0.113 | zgc:110216 | -0.050 |
| elavl3 | 0.111 | akap12b | -0.049 |
| atp6v0cb | 0.110 | gar1 | -0.049 |