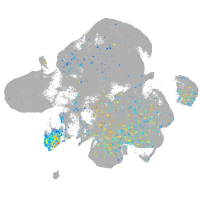
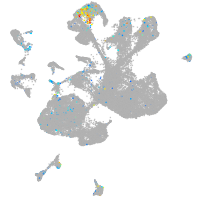
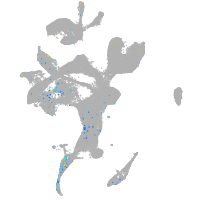
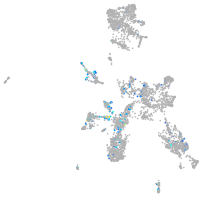
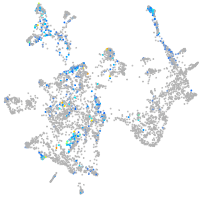
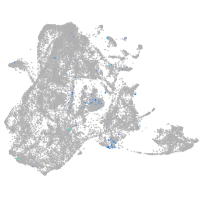
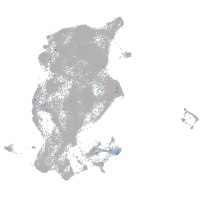
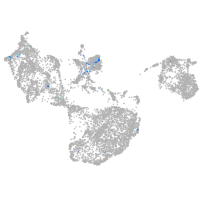
transmembrane protein with EGF-like and two follistatin-like domains 2b
ZFIN 
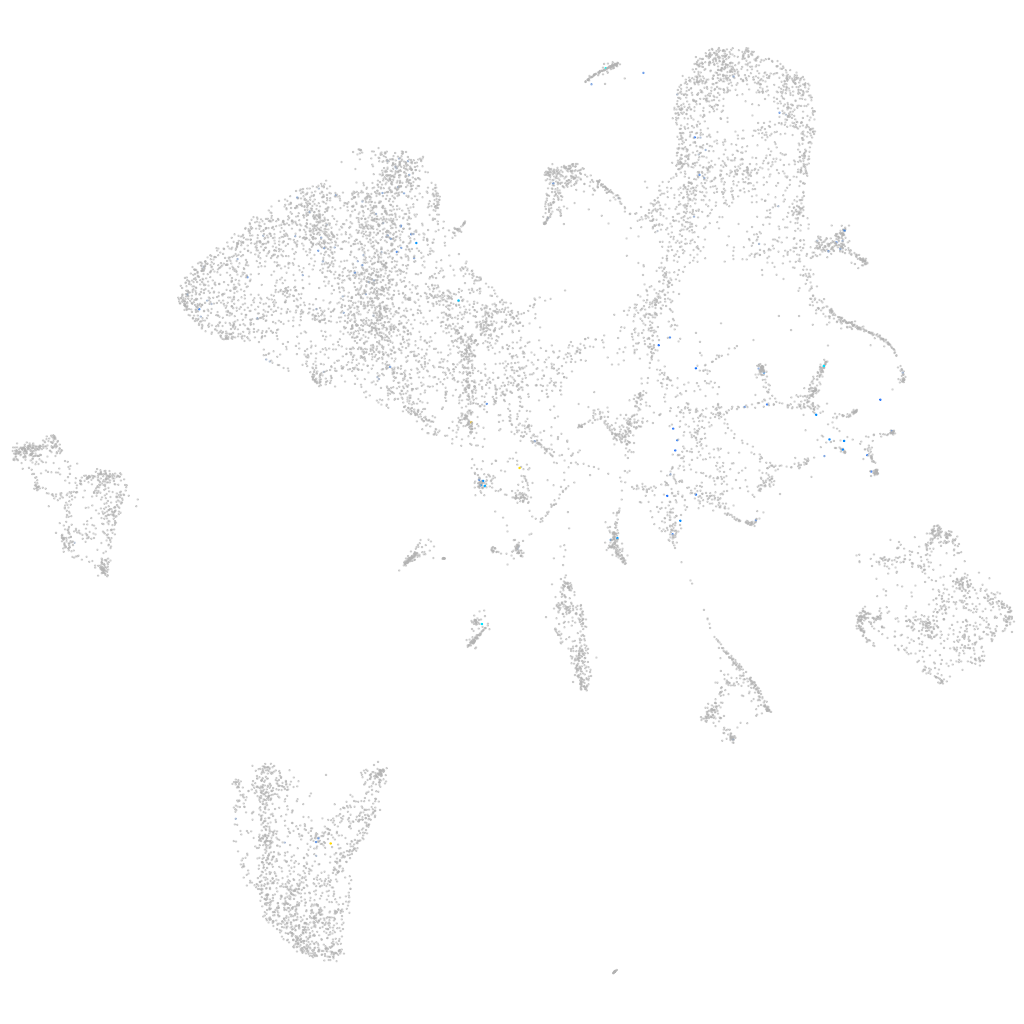
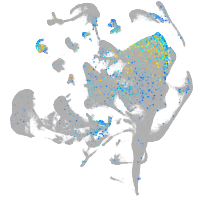
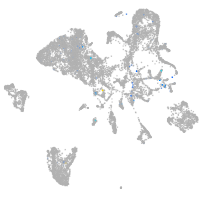
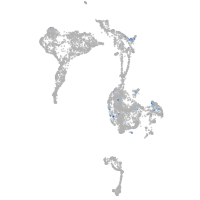
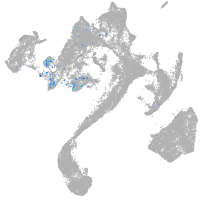
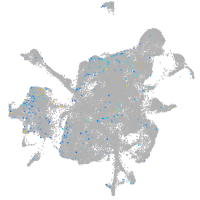
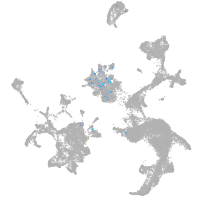
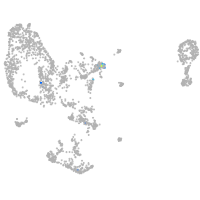
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| BX936415.1 | 0.334 | rpl30 | -0.037 |
| tunar | 0.324 | sec61b | -0.036 |
| XLOC-006820 | 0.320 | rpl6 | -0.035 |
| LOC110439875 | 0.316 | ssr2 | -0.033 |
| lrrc3ca | 0.271 | ssr3 | -0.032 |
| trpc6b | 0.270 | nupr1b | -0.031 |
| gpr88 | 0.259 | ssr1 | -0.030 |
| CABZ01079116.1 | 0.243 | fth1a | -0.030 |
| si:ch73-233f7.6 | 0.237 | apoeb | -0.029 |
| mmp28 | 0.223 | ahcy | -0.029 |
| zgc:153118 | 0.221 | rpl23a | -0.028 |
| si:dkey-261m9.9 | 0.217 | dap | -0.028 |
| XLOC-036573 | 0.216 | fabp2 | -0.028 |
| trim46b | 0.215 | gatm | -0.027 |
| fam155a | 0.211 | tram1 | -0.027 |
| nt5c1aa | 0.209 | sec61a1l | -0.026 |
| myadml2 | 0.209 | atp5fa1 | -0.026 |
| frmpd3 | 0.205 | rrbp1a | -0.026 |
| cort | 0.204 | snu13b | -0.025 |
| desmb | 0.199 | rad23b | -0.025 |
| XLOC-014755 | 0.190 | gamt | -0.025 |
| lingo2a | 0.187 | lman1 | -0.025 |
| kcnq3 | 0.186 | si:ch211-217k17.7 | -0.025 |
| gabrd | 0.185 | eef1da | -0.024 |
| adgrb1a | 0.183 | slc16a10 | -0.024 |
| XLOC-015481 | 0.182 | sil1 | -0.024 |
| zbbx | 0.181 | eef2b | -0.024 |
| LOC101885553 | 0.179 | atp5mc3b | -0.024 |
| prph2b | 0.178 | gapdh | -0.024 |
| cacna1g | 0.178 | sec61g | -0.024 |
| CU861477.1 | 0.177 | mrto4 | -0.024 |
| dnajb5 | 0.176 | cgref1 | -0.024 |
| XLOC-015238 | 0.174 | eif4ebp3l | -0.023 |
| LOC110438360 | 0.172 | COX5B | -0.023 |
| il12bb | 0.171 | si:ch211-105c13.3 | -0.023 |