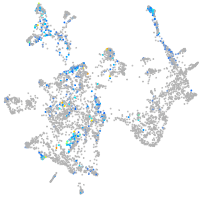
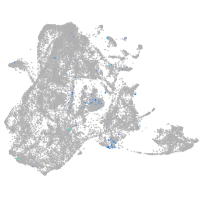
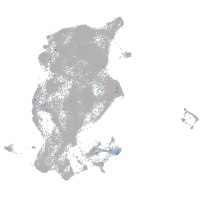
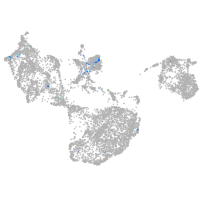
transmembrane protein with EGF-like and two follistatin-like domains 2b
ZFIN 
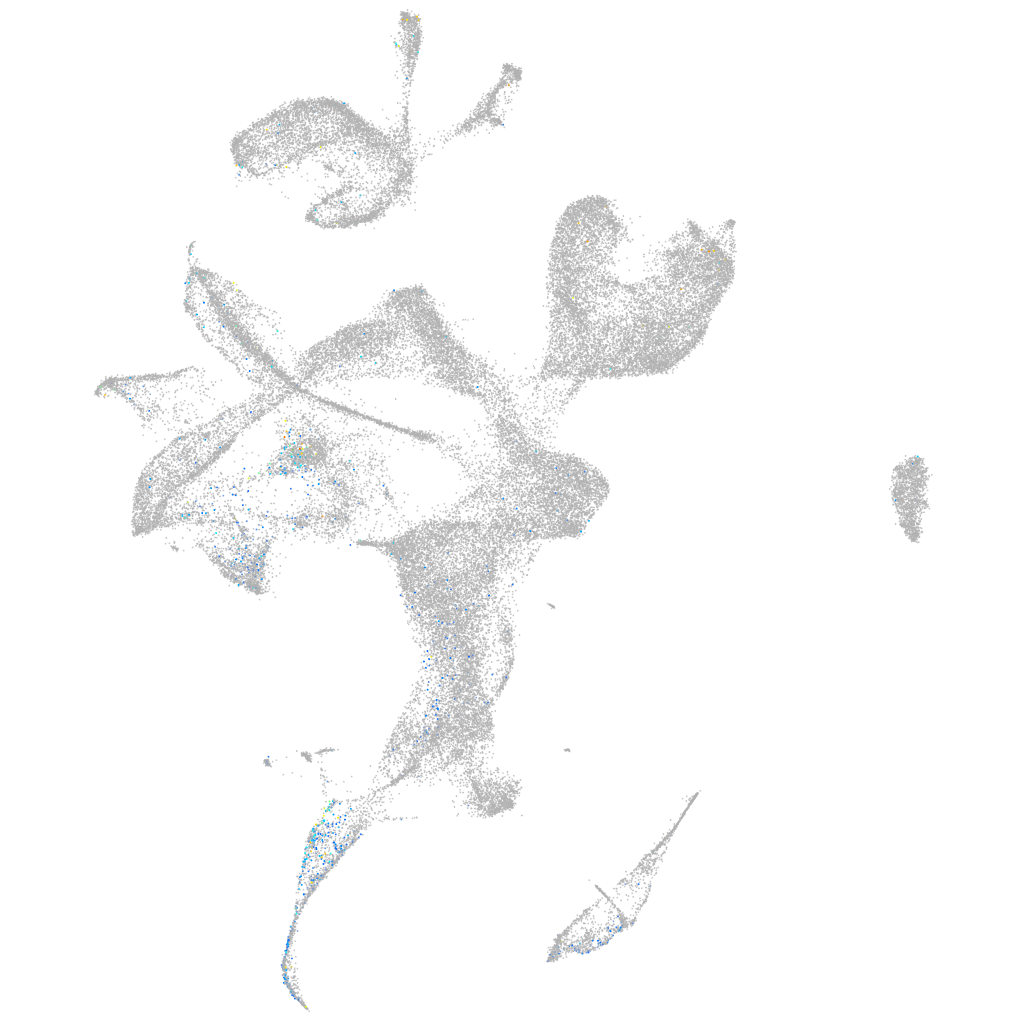
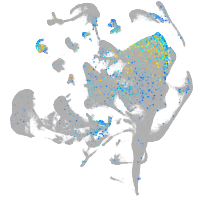
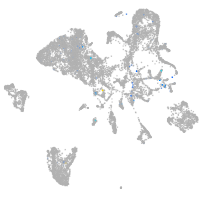
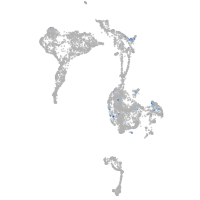
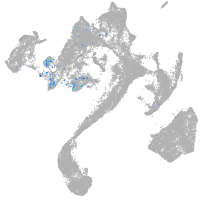
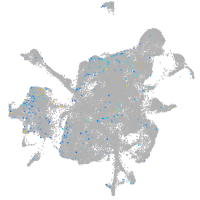
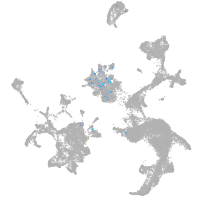
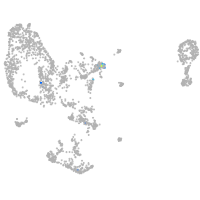
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tyrp1b | 0.180 | ptmab | -0.093 |
| pmela | 0.175 | si:ch73-1a9.3 | -0.066 |
| dct | 0.175 | hmgn6 | -0.057 |
| pmelb | 0.173 | hmgn2 | -0.050 |
| bace2 | 0.167 | hmga1a | -0.050 |
| oca2 | 0.166 | si:ch211-137a8.4 | -0.044 |
| tspan36 | 0.165 | hnrnpaba | -0.044 |
| tyrp1a | 0.164 | hmgb2a | -0.042 |
| slc45a2 | 0.164 | fabp7a | -0.042 |
| comtb | 0.163 | h2afva | -0.039 |
| rab38 | 0.157 | rorb | -0.039 |
| sytl2b | 0.156 | khdrbs1a | -0.038 |
| col4a5 | 0.156 | cirbpb | -0.038 |
| tspan10 | 0.155 | zgc:153409 | -0.037 |
| gpr143 | 0.155 | neurod4 | -0.037 |
| rab27a | 0.154 | anp32e | -0.037 |
| gstp1 | 0.152 | foxg1b | -0.035 |
| cracr2ab | 0.150 | cadm3 | -0.035 |
| sparc | 0.148 | si:ch73-281n10.2 | -0.035 |
| col4a6 | 0.146 | rbbp4 | -0.034 |
| mlphb | 0.145 | hmgb2b | -0.033 |
| tfr1a | 0.143 | si:ch211-222l21.1 | -0.033 |
| sytl2a | 0.142 | h3f3d | -0.033 |
| fstl1b | 0.141 | h2afvb | -0.032 |
| triobpa | 0.140 | hmgb1a | -0.032 |
| abcg2d | 0.136 | fabp3 | -0.032 |
| mitfa | 0.136 | marcksl1b | -0.032 |
| qdpra | 0.136 | epb41a | -0.031 |
| ldlrap1a | 0.135 | nono | -0.031 |
| vat1 | 0.135 | hmgb1b | -0.031 |
| si:dkey-21a6.5 | 0.134 | sinhcafl | -0.031 |
| slc24a5 | 0.134 | crx | -0.031 |
| LOC103910009 | 0.134 | ilf2 | -0.030 |
| FP085398.1 | 0.133 | tp53inp2 | -0.030 |
| zgc:110591 | 0.133 | ndrg1b | -0.030 |