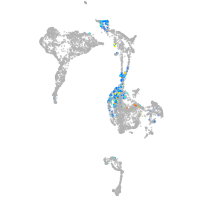
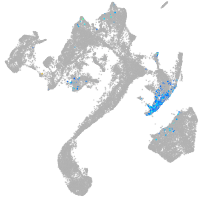
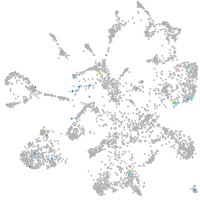
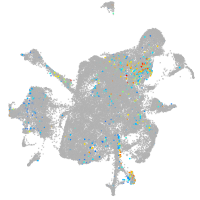
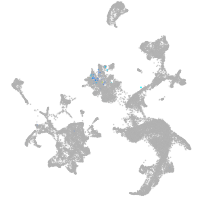
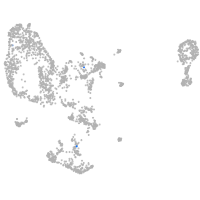
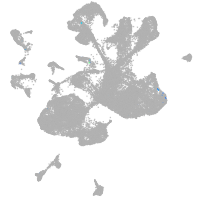
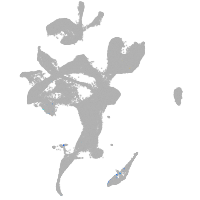
thrombospondin 2a
ZFIN 
Other cell groups


Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dand5 | 0.287 | nova2 | -0.040 |
| LOC101885088 | 0.234 | gpm6aa | -0.036 |
| pttg1ipa | 0.198 | fabp3 | -0.033 |
| pkd1l1 | 0.190 | tuba1c | -0.030 |
| CR450832.1 | 0.180 | marcksl1b | -0.029 |
| wnt2bb | 0.178 | rtn1a | -0.026 |
| FO907087.1 | 0.175 | CU467822.1 | -0.026 |
| cdx1a | 0.172 | elavl3 | -0.026 |
| snai1a | 0.151 | CU634008.1 | -0.024 |
| LOC101886835 | 0.149 | meis1b | -0.024 |
| fgf8a | 0.147 | ckbb | -0.023 |
| tbxta | 0.146 | si:dkey-56m19.5 | -0.023 |
| tll1 | 0.134 | si:dkey-276j7.1 | -0.023 |
| noto | 0.134 | ncam1a | -0.023 |
| cxcr4a | 0.128 | cspg5a | -0.023 |
| wu:fc23c09 | 0.124 | dpysl2b | -0.022 |
| bcl3 | 0.122 | pvalb2 | -0.022 |
| nradd | 0.119 | zgc:165461 | -0.022 |
| sp5l | 0.118 | mdkb | -0.021 |
| fgf3 | 0.118 | tmsb | -0.021 |
| mir196c | 0.113 | gpm6bb | -0.021 |
| CR854964.1 | 0.112 | gfap | -0.021 |
| clul1 | 0.111 | midn | -0.021 |
| BX927327.1 | 0.110 | slc1a2b | -0.020 |
| fgf17 | 0.110 | tuba1a | -0.020 |
| si:ch211-129o18.4 | 0.109 | hbbe1.3 | -0.020 |
| acot16 | 0.106 | si:ch73-21g5.7 | -0.020 |
| fgf4 | 0.105 | hapln1a | -0.019 |
| tbxtb | 0.103 | fabp7a | -0.019 |
| RF01233 | 0.098 | pou3f3b | -0.019 |
| BX649388.1 | 0.097 | atp1a1b | -0.019 |
| apela | 0.096 | actc1b | -0.019 |
| fn1b | 0.094 | gapdhs | -0.019 |
| si:dkey-266f7.5 | 0.093 | hmgb3a | -0.019 |
| taar20j | 0.090 | msi1 | -0.019 |