si:ch211-129o18.4
ZFIN 
Other cell groups


all cells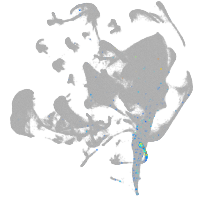 |
blastomeres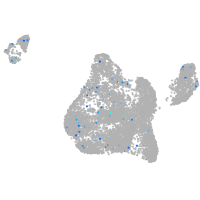 |
PGCs |
endoderm |
axial mesoderm |
muscle 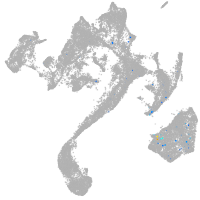 |
mural cells / non-skeletal muscle 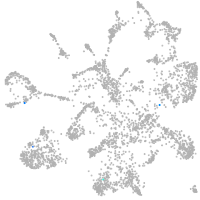 |
mesenchyme  |
hematopoietic / vasculature 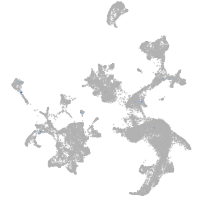 |
pronephros 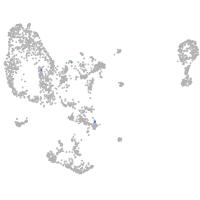 |
neural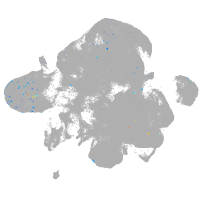 |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis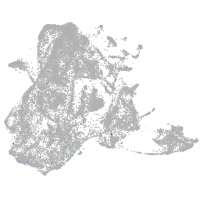 |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-27e6.1 | 0.171 | pvalb1 | -0.027 |
| noto | 0.118 | pvalb2 | -0.026 |
| fgf4 | 0.108 | krt4 | -0.025 |
| col15a1a | 0.095 | cyt1 | -0.024 |
| tbxta | 0.095 | gapdhs | -0.024 |
| emilin3a | 0.089 | actc1b | -0.023 |
| loxl5b | 0.085 | calm1a | -0.023 |
| sst6 | 0.072 | ccni | -0.022 |
| angptl6 | 0.071 | gng3 | -0.022 |
| tbx3b | 0.070 | gpm6aa | -0.022 |
| admp | 0.069 | hbbe1.3 | -0.022 |
| b3gnt7l | 0.069 | icn | -0.022 |
| fgf20a | 0.069 | rtn1a | -0.022 |
| ntd5 | 0.068 | stmn1b | -0.022 |
| fadd | 0.064 | atp6v0cb | -0.021 |
| fgf3 | 0.064 | cyt1l | -0.021 |
| hspb1 | 0.064 | gpm6ab | -0.021 |
| sp5l | 0.064 | hbae3 | -0.021 |
| twist2 | 0.063 | krtt1c19e | -0.021 |
| fgf8a | 0.062 | anxa1a | -0.020 |
| foxa | 0.061 | ckbb | -0.020 |
| foxa2 | 0.061 | rnasekb | -0.020 |
| cdx4 | 0.058 | s100a10b | -0.020 |
| cx43.4 | 0.055 | sncb | -0.020 |
| apela | 0.052 | tmsb1 | -0.020 |
| nop58 | 0.052 | tuba1c | -0.020 |
| dkc1 | 0.050 | elavl3 | -0.019 |
| hjv | 0.050 | elavl4 | -0.019 |
| lrrc59 | 0.049 | hbae1.1 | -0.019 |
| npm1a | 0.049 | icn2 | -0.019 |
| plod2 | 0.049 | krt5 | -0.019 |
| prtfdc1 | 0.048 | si:ch211-195b11.3 | -0.019 |
| fbl | 0.047 | vamp2 | -0.019 |
| fibinb | 0.047 | ywhag2 | -0.019 |
| her7 | 0.047 | atp6v1e1b | -0.018 |




