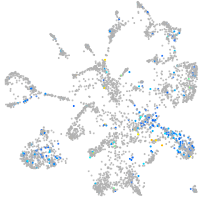
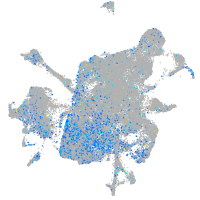
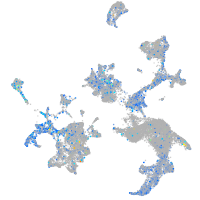
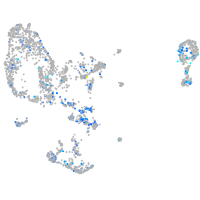
"transcription factor Dp-1, b"
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rrm1 | 0.269 | pvalb1 | -0.129 |
| pcna | 0.266 | pvalb2 | -0.109 |
| cks1b | 0.261 | CR383676.1 | -0.098 |
| dut | 0.257 | tob1b | -0.097 |
| cdca5 | 0.253 | COX3 | -0.097 |
| rpa2 | 0.251 | zgc:158463 | -0.096 |
| ccna2 | 0.249 | eef1da | -0.087 |
| hmgb2a | 0.243 | apoa1b | -0.086 |
| rrm2 | 0.242 | b2ml | -0.086 |
| anp32b | 0.237 | actc1b | -0.084 |
| tubb2b | 0.235 | apoa2 | -0.083 |
| cdk1 | 0.235 | tmem59 | -0.083 |
| seta | 0.235 | mt-co2 | -0.082 |
| slbp | 0.234 | spint2 | -0.080 |
| setb | 0.233 | CELA1 (1 of many) | -0.079 |
| mki67 | 0.233 | rplp2 | -0.079 |
| chaf1a | 0.232 | pvalb8 | -0.076 |
| mad2l1 | 0.231 | col9a1a | -0.075 |
| smc2 | 0.228 | atp2b1a | -0.075 |
| stmn1a | 0.226 | cyp2ad3 | -0.075 |
| banf1 | 0.226 | slc3a2b | -0.074 |
| sumo3b | 0.226 | nupr1a | -0.074 |
| snrpd1 | 0.225 | prss1 | -0.074 |
| rbbp4 | 0.225 | hbp1 | -0.072 |
| aurkb | 0.224 | cebpd | -0.072 |
| dek | 0.224 | atp6v1aa | -0.071 |
| rpa3 | 0.223 | ip6k2a | -0.070 |
| ranbp1 | 0.223 | ckbb | -0.070 |
| hmga1a | 0.223 | krt17 | -0.069 |
| dhfr | 0.222 | dedd1 | -0.069 |
| mibp | 0.222 | prss59.2 | -0.068 |
| nasp | 0.221 | gabarapl2 | -0.068 |
| zgc:110216 | 0.221 | h1f0 | -0.068 |
| ncapg | 0.221 | gngt2b | -0.068 |
| lbr | 0.220 | gapdhs | -0.067 |