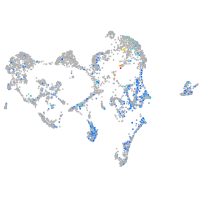
"transcription factor Dp-1, b"
ZFIN 
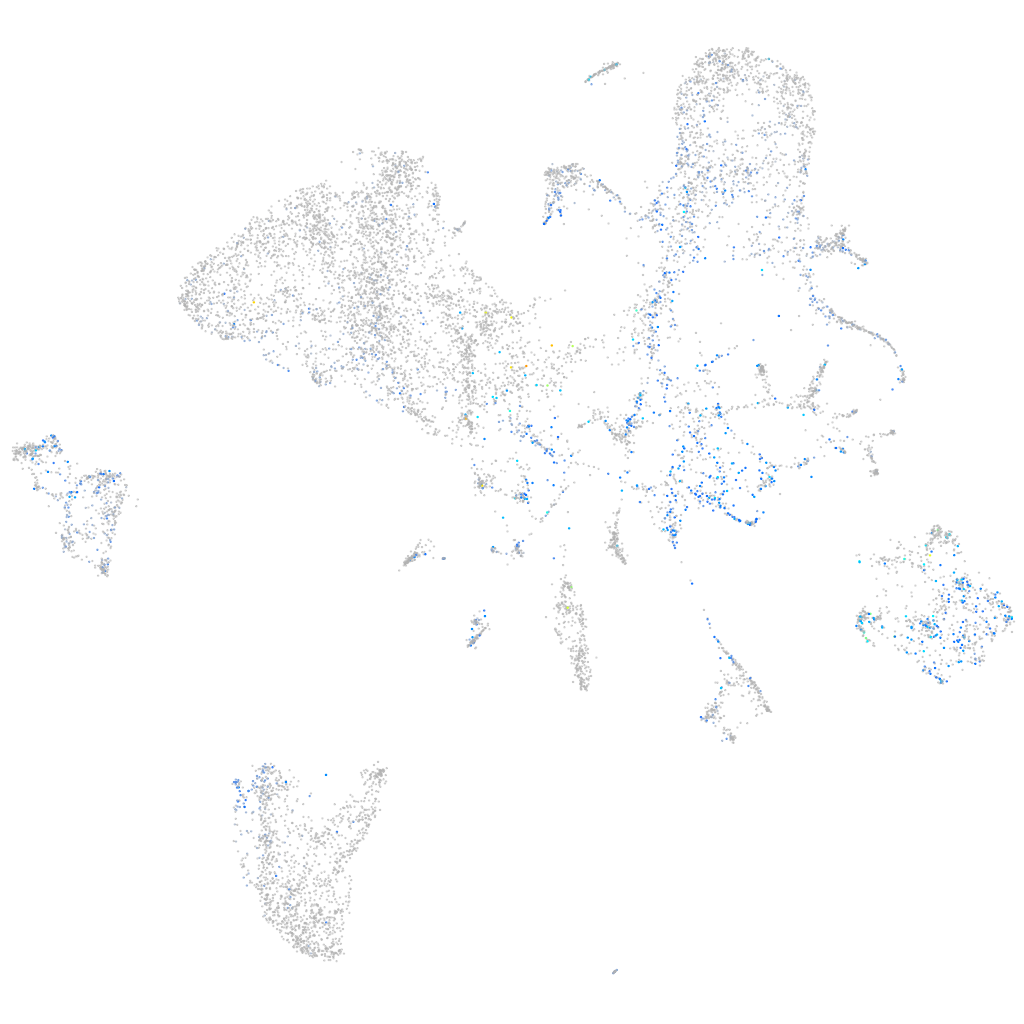
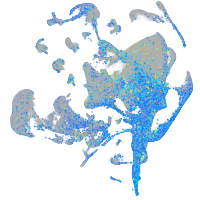
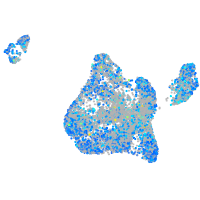
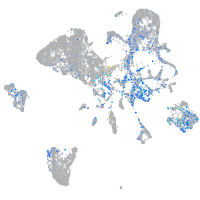
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hmgb2a | 0.296 | gamt | -0.205 |
| hmgb2b | 0.288 | gatm | -0.201 |
| hmga1a | 0.285 | gapdh | -0.194 |
| stmn1a | 0.284 | bhmt | -0.184 |
| si:ch211-222l21.1 | 0.284 | pnp4b | -0.179 |
| tuba8l4 | 0.281 | agxtb | -0.170 |
| hnrnpaba | 0.281 | apoa1b | -0.170 |
| h2afvb | 0.276 | apoa2 | -0.168 |
| khdrbs1a | 0.274 | fbp1b | -0.165 |
| tubb2b | 0.274 | apoa4b.1 | -0.164 |
| pcna | 0.272 | gpx4a | -0.163 |
| seta | 0.271 | mat1a | -0.161 |
| cirbpa | 0.269 | abat | -0.159 |
| cbx3a | 0.268 | rbp4 | -0.158 |
| si:ch73-281n10.2 | 0.267 | afp4 | -0.158 |
| setb | 0.266 | fabp10a | -0.155 |
| anp32b | 0.266 | zgc:123103 | -0.155 |
| cbx5 | 0.264 | ces2 | -0.154 |
| hnrnpa0b | 0.264 | grhprb | -0.154 |
| h3f3d | 0.262 | apom | -0.152 |
| syncrip | 0.262 | nupr1b | -0.151 |
| hmgn2 | 0.261 | mgst1.2 | -0.151 |
| hnrnpabb | 0.259 | glud1b | -0.150 |
| h2afva | 0.258 | ahcy | -0.150 |
| sumo3a | 0.258 | fetub | -0.150 |
| snrpd1 | 0.258 | hao1 | -0.150 |
| chaf1a | 0.256 | serpina1 | -0.149 |
| snrpb | 0.255 | apobb.1 | -0.149 |
| snrpf | 0.253 | zgc:92744 | -0.149 |
| si:ch73-1a9.3 | 0.252 | tfa | -0.149 |
| anp32a | 0.252 | gc | -0.148 |
| hdac1 | 0.252 | ttr | -0.148 |
| nasp | 0.251 | LOC110437731 | -0.147 |
| ptmab | 0.251 | ckba | -0.147 |
| sumo3b | 0.251 | pklr | -0.147 |