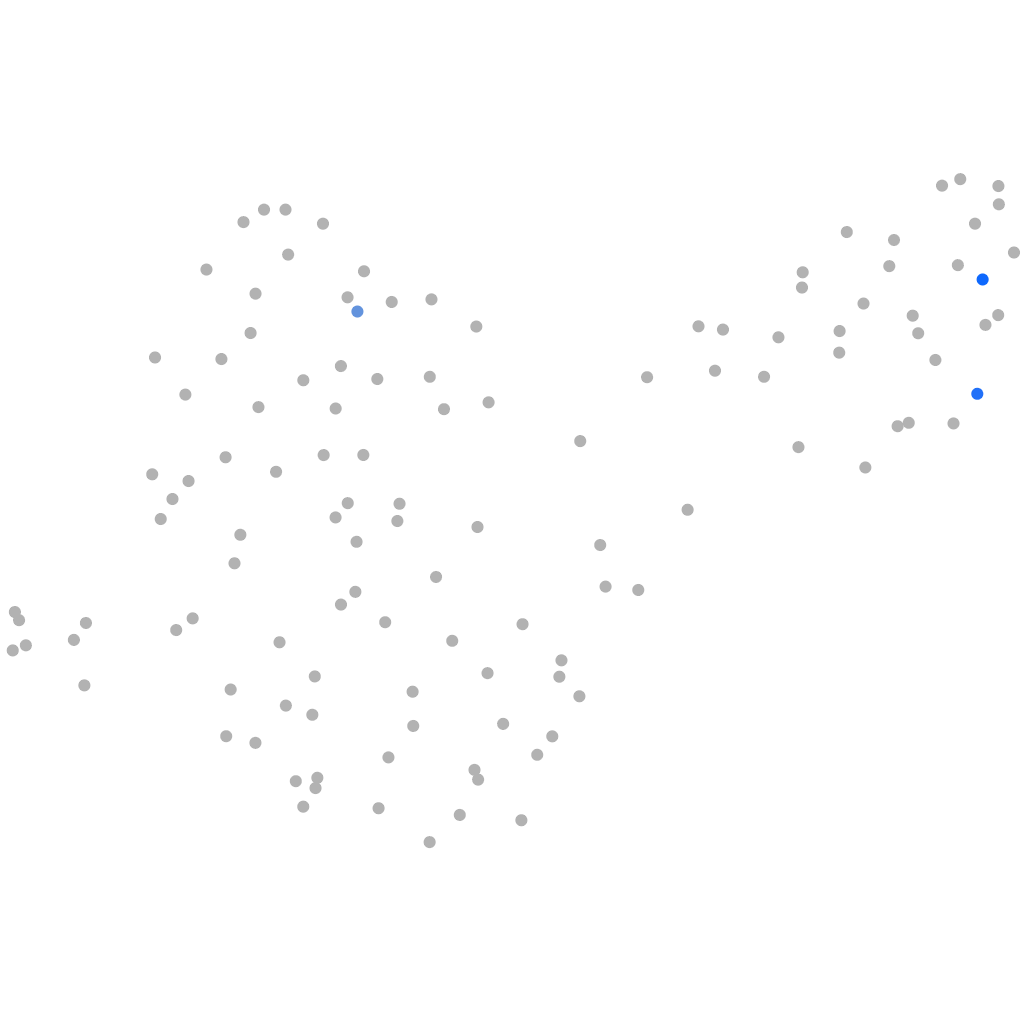
transcription factor CP2-like 1
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm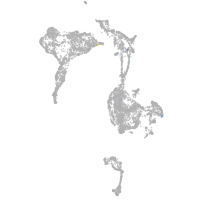 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme 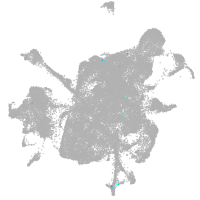 |
hematopoietic / vasculature 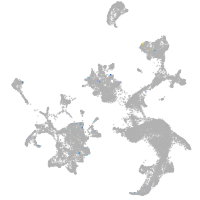 |
pronephros 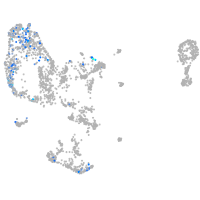 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| amigo1 | 0.921 | smarca4a | -0.183 |
| iqsec2a | 0.921 | si:dkey-67c22.2 | -0.179 |
| CR759845.1 | 0.825 | ncl | -0.175 |
| si:dkey-16p21.7 | 0.774 | wasf2 | -0.160 |
| si:ch211-201h21.5 | 0.774 | smndc1 | -0.158 |
| fads2 | 0.767 | ddx18 | -0.154 |
| sema6d | 0.750 | dhx16 | -0.151 |
| tti1 | 0.739 | h3f3d | -0.149 |
| fam78ab | 0.733 | sltm | -0.149 |
| tbxas1 | 0.696 | gnl3 | -0.147 |
| sgsm1a | 0.672 | sf3b2 | -0.143 |
| XLOC-036690 | 0.672 | sec62 | -0.141 |
| atp2a2a | 0.672 | rcc1 | -0.140 |
| BX510960.2 | 0.672 | tbx16 | -0.139 |
| c4 | 0.672 | stm | -0.139 |
| ccdc17 | 0.672 | sec63 | -0.135 |
| CR377210.1 | 0.672 | npm1a | -0.135 |
| CR626887.2 | 0.672 | szrd1 | -0.135 |
| CT583646.2 | 0.672 | ebna1bp2 | -0.135 |
| CU914550.1 | 0.672 | nhp2 | -0.134 |
| dhx32b | 0.672 | tra2b | -0.134 |
| fam43a | 0.672 | slc16a3 | -0.133 |
| FO704745.1 | 0.672 | zgc:56676 | -0.133 |
| gad2 | 0.672 | surf2 | -0.131 |
| igsf5a | 0.672 | NC-002333.4 | -0.131 |
| irf8 | 0.672 | mfap1 | -0.130 |
| klf5a | 0.672 | lyar | -0.128 |
| LOC100331291 | 0.672 | ddit4 | -0.128 |
| LOC100332187 | 0.672 | cdc40 | -0.127 |
| LOC101884404 | 0.672 | golga7 | -0.127 |
| LOC103911115 | 0.672 | asb11 | -0.125 |
| LOC110439449 | 0.672 | bnip4 | -0.124 |
| myct1b | 0.672 | ammecr1 | -0.123 |
| phox2ba | 0.672 | dynll2a | -0.123 |
| si:ch211-285c6.6 | 0.672 | rbm38 | -0.123 |