transcription factor CP2-like 1
ZFIN 
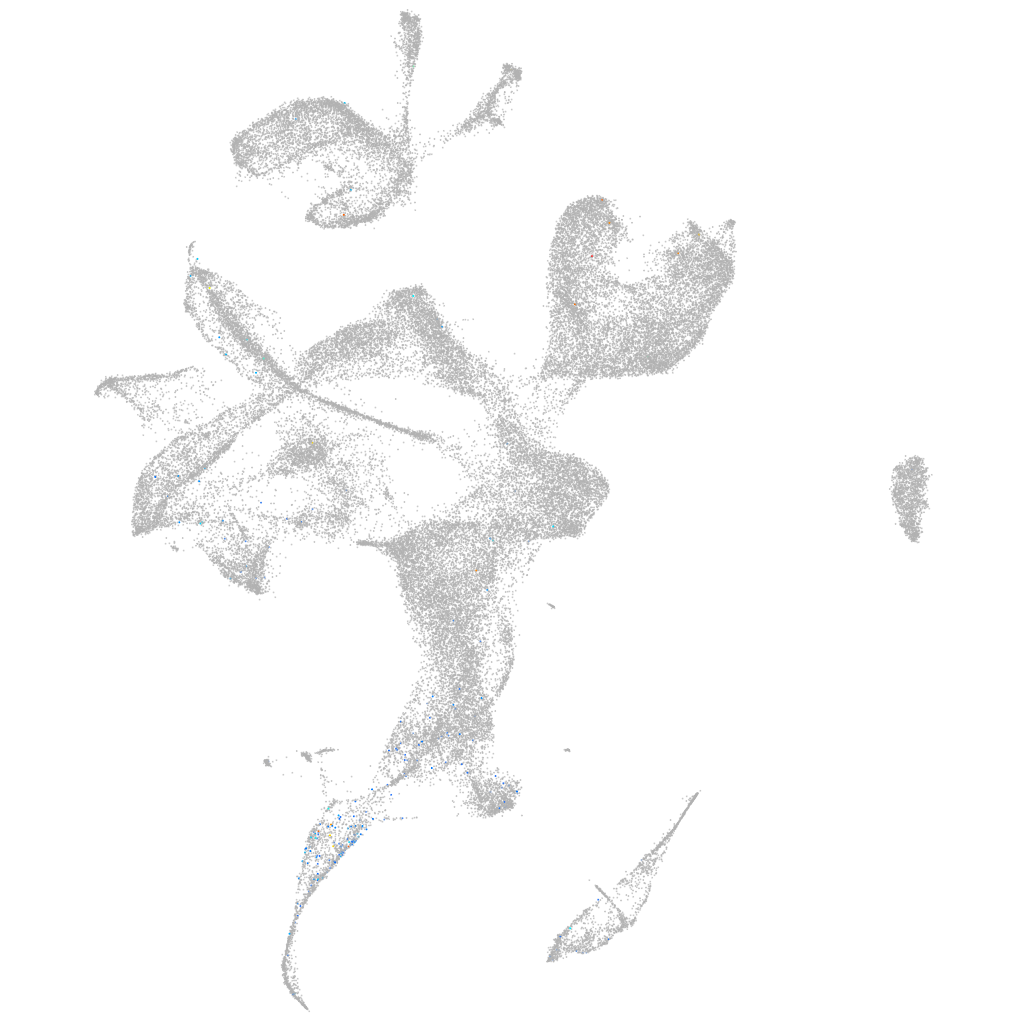
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm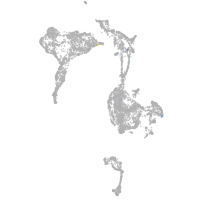 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme 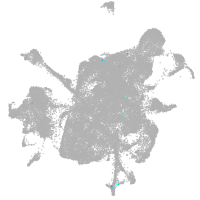 |
hematopoietic / vasculature 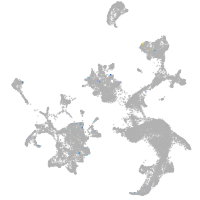 |
pronephros 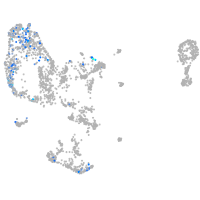 |
neural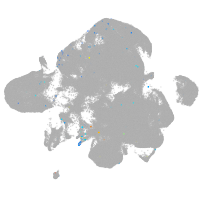 |
spinal cord / glia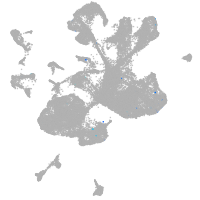 |
eye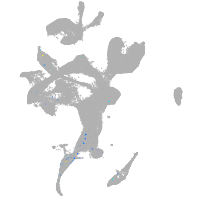 |
taste / olfactory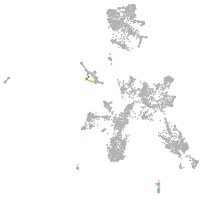 |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tmem88b | 0.131 | ptmab | -0.048 |
| slc45a2 | 0.123 | hmgb1a | -0.033 |
| tfec | 0.117 | ptmaa | -0.032 |
| FP085398.1 | 0.115 | tuba1c | -0.031 |
| cax1 | 0.112 | gpm6aa | -0.031 |
| dct | 0.112 | fabp7a | -0.029 |
| agtrap | 0.111 | btg1 | -0.029 |
| sytl2b | 0.111 | si:ch211-137a8.4 | -0.028 |
| wnt2 | 0.110 | cadm3 | -0.028 |
| zgc:110591 | 0.108 | si:ch73-1a9.3 | -0.027 |
| LOC101883894 | 0.107 | anp32e | -0.027 |
| pmela | 0.105 | gpm6ab | -0.026 |
| BX470229.1 | 0.105 | hmgn6 | -0.026 |
| tspan36 | 0.104 | ckbb | -0.026 |
| tyrp1a | 0.104 | rorb | -0.026 |
| mitfa | 0.103 | hnrnpaba | -0.025 |
| rab32a | 0.103 | tuba1a | -0.024 |
| gpr143 | 0.102 | hmgb3a | -0.023 |
| tyrp1b | 0.101 | hes6 | -0.022 |
| pcbd1 | 0.098 | h2afva | -0.022 |
| pah | 0.097 | mdkb | -0.021 |
| qdpra | 0.096 | marcksl1b | -0.021 |
| slc24a5 | 0.096 | neurod4 | -0.020 |
| fabp11a | 0.095 | tp53inp2 | -0.020 |
| si:dkey-225f5.5 | 0.094 | foxg1b | -0.020 |
| FP085399.2 | 0.094 | insm1a | -0.020 |
| gstp1 | 0.094 | cirbpb | -0.020 |
| tyr | 0.094 | hmgb1b | -0.020 |
| zgc:110239 | 0.092 | myt1a | -0.020 |
| alkal2a | 0.092 | hmgn2 | -0.020 |
| trpm1b | 0.090 | scrt2 | -0.019 |
| pmelb | 0.089 | tmem35 | -0.019 |
| rab38 | 0.089 | cspg5a | -0.019 |
| cx43 | 0.087 | mex3b | -0.019 |
| oca2 | 0.087 | ankrd12 | -0.018 |