testis derived transcript (3 LIM domains)
ZFIN 
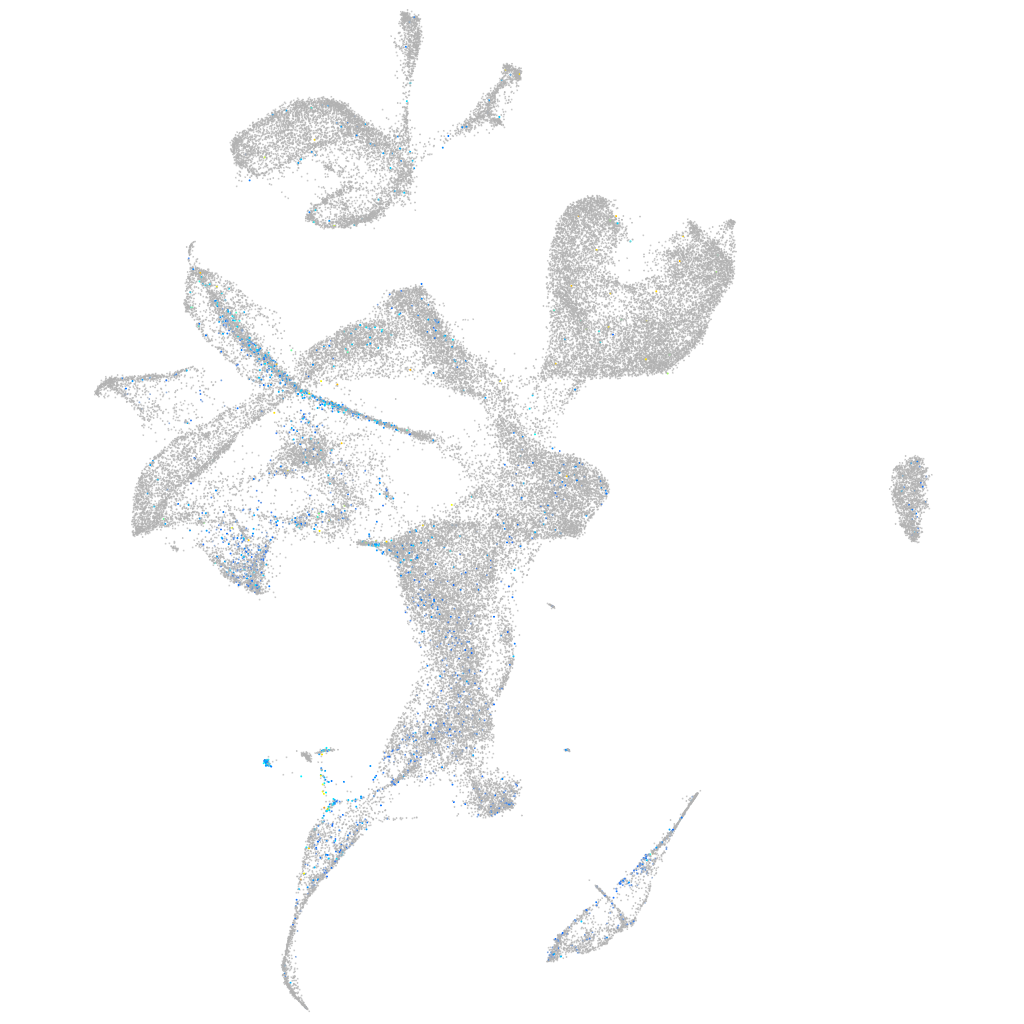
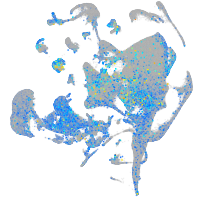
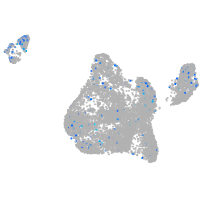
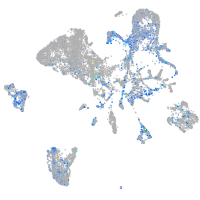
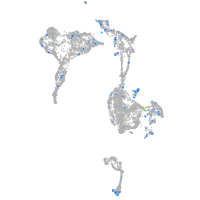
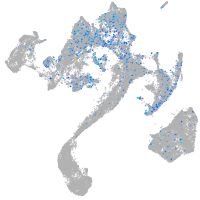
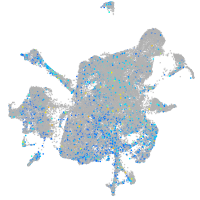
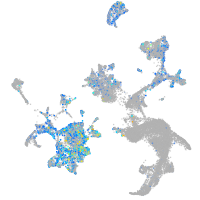
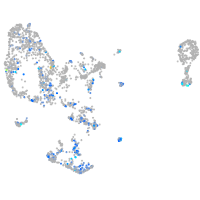
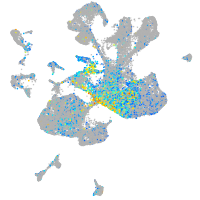
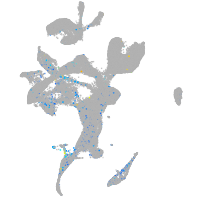
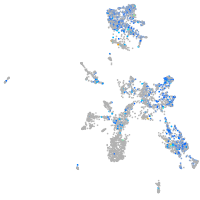
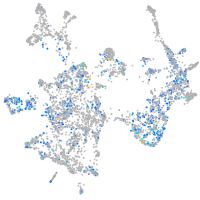
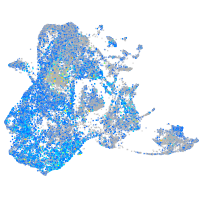
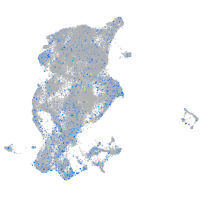
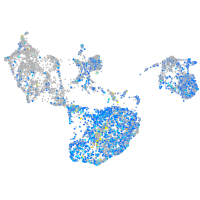
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ednrab | 0.196 | si:ch73-1a9.3 | -0.067 |
| crestin | 0.175 | anp32e | -0.063 |
| alcama | 0.157 | neurod4 | -0.057 |
| rbpms2b | 0.136 | syt5b | -0.057 |
| vim | 0.128 | ptmab | -0.054 |
| tfec | 0.120 | vsx1 | -0.052 |
| msx1b | 0.114 | snap25b | -0.051 |
| adcyap1b | 0.113 | calb2b | -0.050 |
| tpm4a | 0.109 | cadm3 | -0.047 |
| LOC110438542 | 0.105 | crx | -0.047 |
| slc1a3a | 0.105 | rs1a | -0.046 |
| BX248318.1 | 0.104 | nrn1lb | -0.045 |
| zgc:158291 | 0.103 | gnb3a | -0.045 |
| wls | 0.103 | pcp4l1 | -0.045 |
| XLOC-001964 | 0.099 | sypb | -0.045 |
| cnn2 | 0.099 | samsn1a | -0.044 |
| vat1 | 0.098 | hmgb2a | -0.044 |
| slc15a2 | 0.095 | ndrg1b | -0.043 |
| sox10 | 0.095 | otx5 | -0.043 |
| tmsb | 0.094 | bhlhe23 | -0.042 |
| mllt11 | 0.092 | vamp1 | -0.041 |
| barhl1a | 0.092 | lin7a | -0.041 |
| islr2 | 0.091 | histh1l | -0.041 |
| inka1a | 0.091 | cnrip1b | -0.041 |
| cldn7a | 0.088 | pcbp3 | -0.040 |
| pmp22a | 0.088 | ndrg4 | -0.040 |
| hspg2 | 0.088 | scg3 | -0.039 |
| si:ch211-11c3.9 | 0.088 | ppp1r1b | -0.039 |
| fstl1b | 0.087 | cplx4a | -0.038 |
| rbpms2a | 0.085 | mpp6b | -0.038 |
| tspan36 | 0.085 | si:ch1073-83n3.2 | -0.037 |
| sparc | 0.084 | atp6v0cb | -0.037 |
| stmn2b | 0.084 | cspg5b | -0.036 |
| sdc4 | 0.084 | stx3a | -0.035 |
| eva1ba | 0.084 | BX663503.1 | -0.035 |