"transcription elongation factor A (SII), 3"
ZFIN 
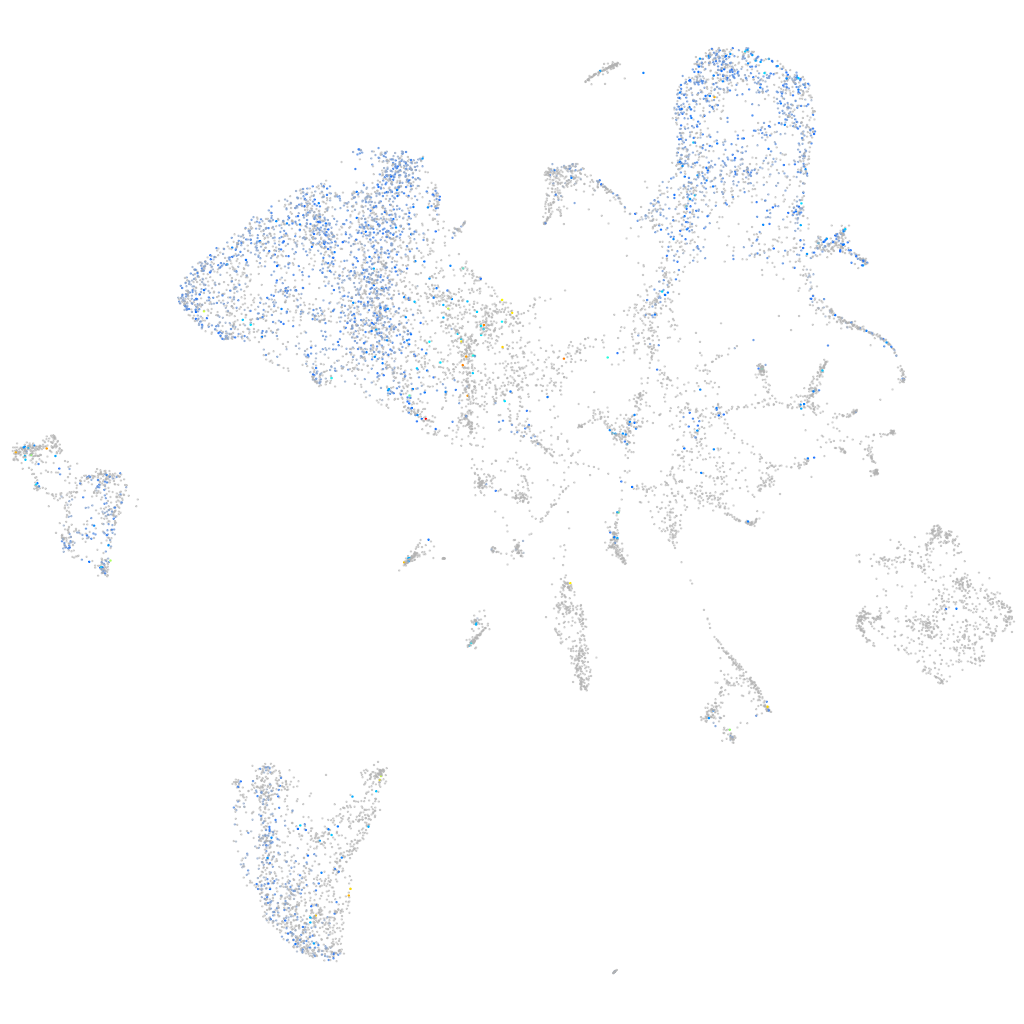
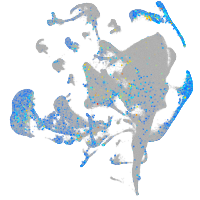
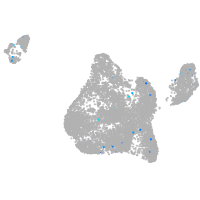
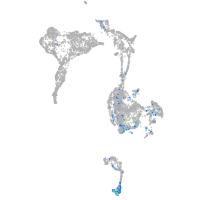
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| glud1b | 0.237 | si:ch211-222l21.1 | -0.176 |
| sult2st2 | 0.232 | hmgn6 | -0.174 |
| scp2a | 0.228 | hmgb3a | -0.164 |
| rdh1 | 0.226 | si:ch73-1a9.3 | -0.161 |
| ugt1a7 | 0.225 | marcksl1b | -0.155 |
| gapdh | 0.225 | hmgb2a | -0.154 |
| mdh1aa | 0.225 | hmgb1b | -0.153 |
| dhrs9 | 0.222 | h2afvb | -0.152 |
| gcshb | 0.221 | marcksb | -0.152 |
| eno3 | 0.219 | nucks1a | -0.151 |
| sod1 | 0.218 | si:ch73-281n10.2 | -0.149 |
| cyp4v8 | 0.216 | hmga1a | -0.148 |
| apoa4b.1 | 0.215 | hnrnpaba | -0.147 |
| cyp8b1 | 0.215 | hmgn2 | -0.146 |
| srd5a2a | 0.215 | khdrbs1a | -0.145 |
| ugt5b4 | 0.214 | h3f3d | -0.144 |
| gstr | 0.212 | ptmab | -0.144 |
| g6pca.2 | 0.211 | hmgb2b | -0.143 |
| gpx4a | 0.210 | tubb2b | -0.142 |
| pgk1 | 0.209 | tuba8l4 | -0.141 |
| dgat2 | 0.208 | syncrip | -0.141 |
| acadl | 0.208 | cx43.4 | -0.141 |
| suclg1 | 0.208 | hdac1 | -0.139 |
| aldob | 0.207 | cirbpb | -0.139 |
| cat | 0.206 | sumo3a | -0.138 |
| agmo | 0.205 | seta | -0.138 |
| fbp1b | 0.205 | hnrnpa0b | -0.137 |
| etnppl | 0.204 | anp32b | -0.136 |
| sdr16c5b | 0.203 | hspb1 | -0.135 |
| rgn | 0.203 | hnrnpa0a | -0.134 |
| sult3st2 | 0.203 | nono | -0.133 |
| haao | 0.202 | stmn1a | -0.133 |
| aldh6a1 | 0.202 | anp32a | -0.133 |
| slc37a4a | 0.202 | akap12b | -0.132 |
| sult1st6 | 0.201 | hnrnpa1a | -0.132 |