T-box transcription factor 2b
ZFIN 
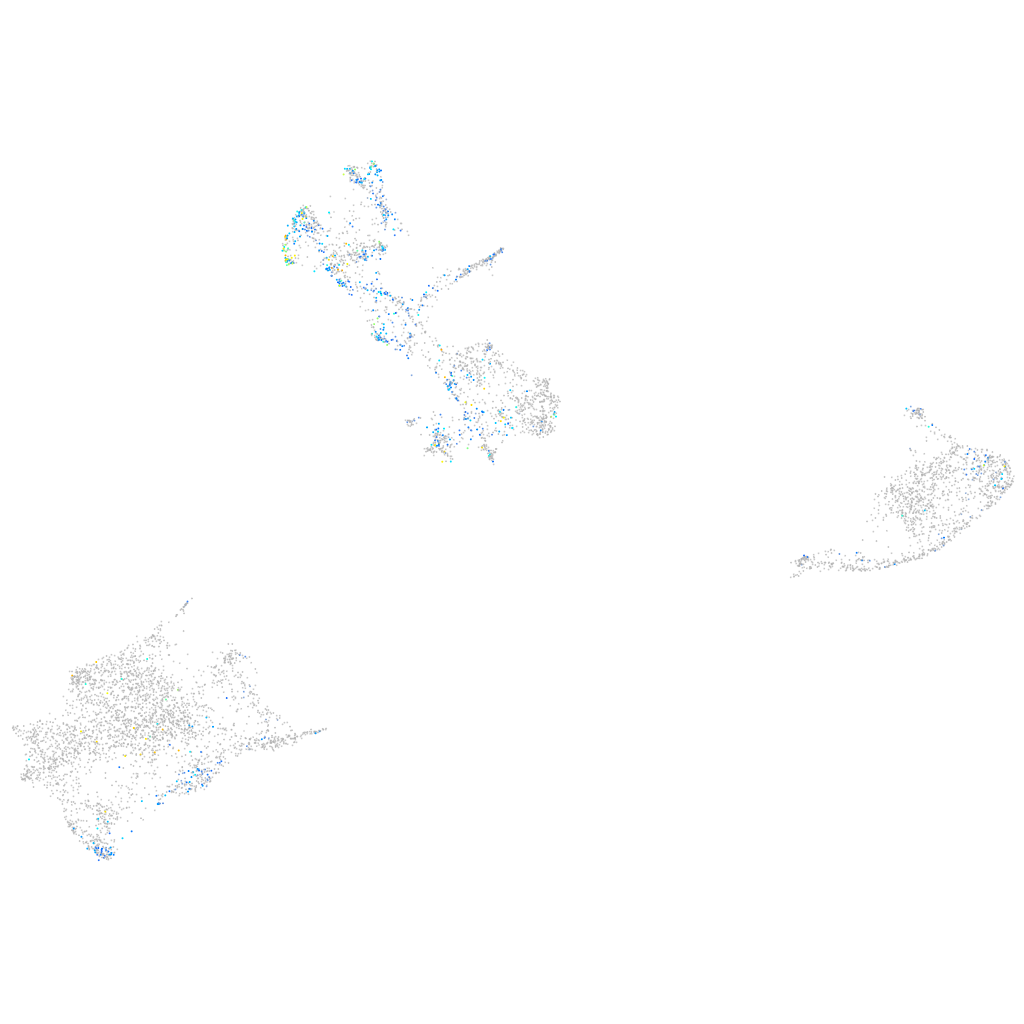
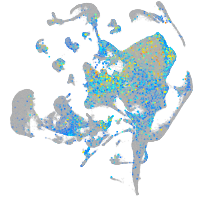
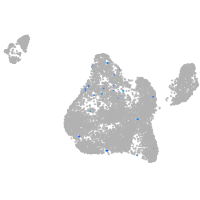
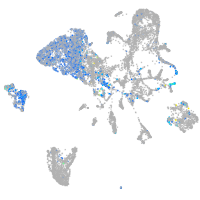
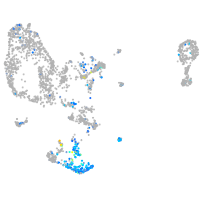
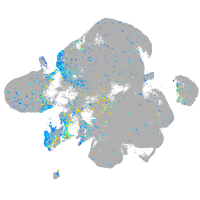
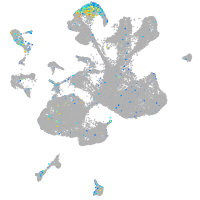
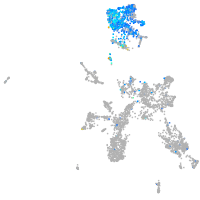
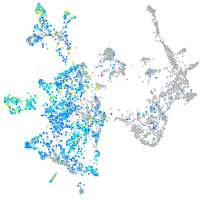
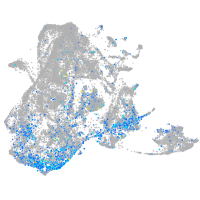
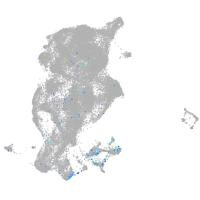
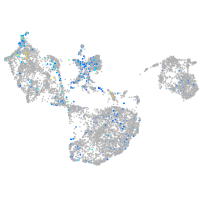
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| alx4a | 0.295 | gch2 | -0.208 |
| si:dkey-197i20.6 | 0.293 | akr1b1 | -0.154 |
| apoda.1 | 0.293 | pts | -0.152 |
| sytl2b | 0.285 | CABZ01021592.1 | -0.149 |
| gpnmb | 0.285 | slc22a7a | -0.148 |
| lypc | 0.279 | cotl1 | -0.140 |
| akap12a | 0.275 | si:dkey-251i10.2 | -0.134 |
| si:ch211-256m1.8 | 0.273 | pcbd1 | -0.131 |
| si:ch73-89b15.3 | 0.270 | fabp3 | -0.121 |
| defbl1 | 0.267 | actb2 | -0.121 |
| alx4b | 0.267 | marcksl1b | -0.119 |
| tcirg1a | 0.264 | cd63 | -0.117 |
| csf1a | 0.259 | zgc:110339 | -0.115 |
| cdh11 | 0.257 | gpd1b | -0.111 |
| ltk | 0.254 | rtn1b | -0.110 |
| alx1 | 0.253 | si:dkey-56m19.5 | -0.109 |
| slc25a36a | 0.251 | icn | -0.106 |
| bambia | 0.250 | qdpra | -0.106 |
| akap12b | 0.250 | tmem243a | -0.105 |
| prx | 0.249 | gstt1a | -0.102 |
| guk1a | 0.248 | tmem130 | -0.098 |
| ppfibp1a | 0.245 | pmela | -0.097 |
| ponzr1 | 0.245 | s100a10b | -0.097 |
| si:ch211-243a20.3 | 0.245 | gstp1 | -0.097 |
| zgc:110789 | 0.244 | mitfa | -0.095 |
| fhl3a | 0.244 | zgc:91968 | -0.093 |
| AL954322.2 | 0.244 | tyrp1b | -0.093 |
| cx43 | 0.236 | anxa1a | -0.091 |
| sytl2a | 0.234 | tfap2e | -0.090 |
| sdc2 | 0.234 | aqp3a | -0.089 |
| pltp | 0.230 | C12orf75 | -0.089 |
| pnp4a | 0.228 | tyrp1a | -0.087 |
| sort1a | 0.226 | gyg1b | -0.086 |
| tpd52l1 | 0.225 | tyr | -0.084 |
| psat1 | 0.225 | prdx1 | -0.082 |