TatD DNase domain containing 1
ZFIN 
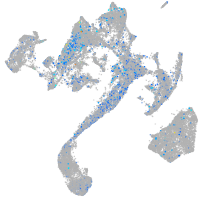
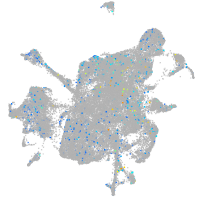
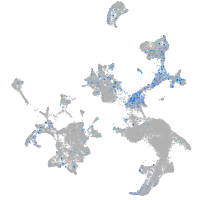
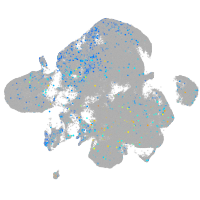
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| npm1a | 0.140 | ptmab | -0.123 |
| nop10 | 0.132 | hmgn6 | -0.081 |
| snu13b | 0.131 | ckbb | -0.079 |
| fabp11a | 0.130 | si:ch73-1a9.3 | -0.078 |
| eif4ebp3l | 0.129 | ptmaa | -0.074 |
| si:dkey-102m7.3 | 0.127 | neurod4 | -0.072 |
| nhp2 | 0.126 | gpm6aa | -0.072 |
| aamp | 0.122 | btg1 | -0.071 |
| rcl1 | 0.122 | hmgb1a | -0.071 |
| paics | 0.121 | epb41a | -0.066 |
| c1qbp | 0.120 | gpm6ab | -0.065 |
| nop58 | 0.120 | vsx1 | -0.063 |
| rps27l | 0.119 | CR383676.1 | -0.063 |
| pa2g4a | 0.118 | tp53inp2 | -0.061 |
| dkc1 | 0.118 | foxg1b | -0.058 |
| tomm20a | 0.116 | cadm3 | -0.057 |
| prps1a | 0.115 | nova2 | -0.055 |
| eif6 | 0.115 | h3f3c | -0.055 |
| bysl | 0.114 | mpp6b | -0.054 |
| nip7 | 0.113 | marcksl1b | -0.054 |
| rrp1 | 0.113 | anp32e | -0.054 |
| adi1 | 0.112 | scrt2 | -0.053 |
| cad | 0.111 | crx | -0.053 |
| nop56 | 0.111 | insm1a | -0.053 |
| ssb | 0.110 | syt5b | -0.052 |
| mrto4 | 0.110 | pvalb1 | -0.050 |
| hspd1 | 0.110 | bcl2l10 | -0.050 |
| gart | 0.110 | CU634008.1 | -0.050 |
| nifk | 0.110 | gng13b | -0.050 |
| bxdc2 | 0.109 | pvalb2 | -0.050 |
| ftsj3 | 0.109 | histh1l | -0.050 |
| fbl | 0.109 | hbbe1.3 | -0.048 |
| rsl1d1 | 0.109 | golga7ba | -0.048 |
| atic | 0.109 | rorb | -0.048 |
| riox2 | 0.108 | actc1b | -0.047 |