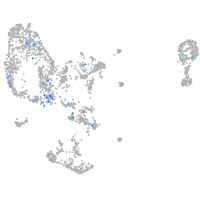
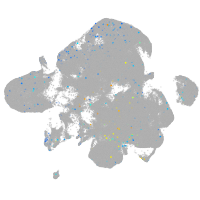
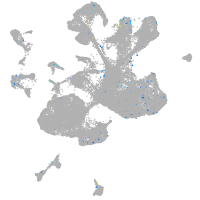
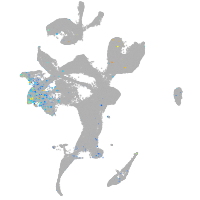
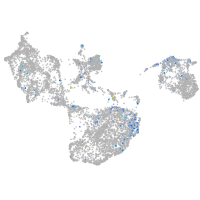
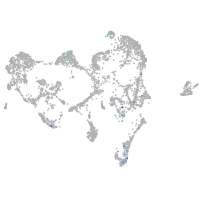
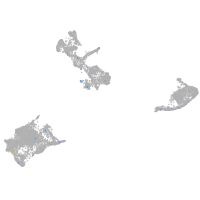
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ptn | 0.240 | gapdh | -0.110 |
| SLC5A10 | 0.235 | ahcy | -0.110 |
| avp | 0.233 | gamt | -0.102 |
| CABZ01053748.1 | 0.213 | gatm | -0.093 |
| ucn3l | 0.206 | bhmt | -0.088 |
| BX005056.1 | 0.199 | fbp1b | -0.088 |
| slc30a2 | 0.195 | gstt1a | -0.087 |
| sst1.1 | 0.192 | mat1a | -0.086 |
| scgn | 0.190 | apoc2 | -0.086 |
| lcp1 | 0.190 | apoa1b | -0.085 |
| cd164l2 | 0.190 | glud1b | -0.084 |
| hivep2b | 0.187 | apoa4b.1 | -0.083 |
| ndufa4l2a | 0.185 | aldh6a1 | -0.082 |
| zgc:175177 | 0.183 | gcshb | -0.081 |
| FO704607.1 | 0.182 | aldh7a1 | -0.080 |
| pcsk2 | 0.182 | afp4 | -0.080 |
| dkk3b | 0.182 | agxta | -0.080 |
| uts1 | 0.175 | aldob | -0.080 |
| apbb3 | 0.173 | scp2a | -0.080 |
| camk1da | 0.172 | eno3 | -0.079 |
| si:dkey-153k10.9 | 0.172 | agxtb | -0.078 |
| pcsk1 | 0.172 | grhprb | -0.077 |
| vat1 | 0.171 | apoa2 | -0.077 |
| LOC110438375 | 0.171 | nipsnap3a | -0.076 |
| CR354610.2 | 0.170 | tfa | -0.076 |
| cntnap2a | 0.169 | cx32.3 | -0.075 |
| elfn1a | 0.169 | slco1d1 | -0.075 |
| kcnj11 | 0.169 | ftcd | -0.075 |
| ins | 0.168 | aqp12 | -0.074 |
| pcdh1a3 | 0.165 | dap | -0.074 |
| LOC100537384 | 0.164 | pnp4b | -0.074 |
| scg2a | 0.163 | suclg1 | -0.073 |
| lrrc4bb | 0.163 | nupr1b | -0.073 |
| neurod1 | 0.163 | msrb2 | -0.073 |
| XLOC-016518 | 0.163 | gstr | -0.073 |