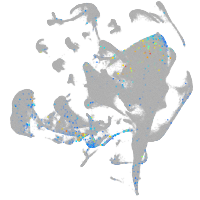
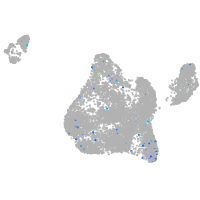
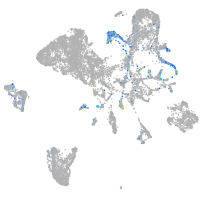
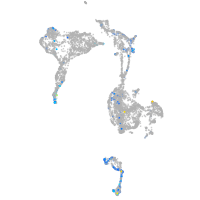
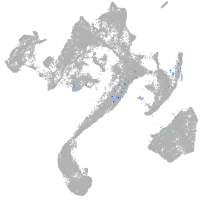
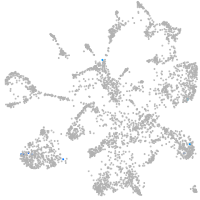
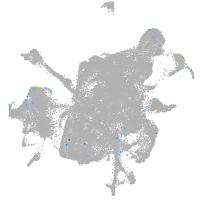
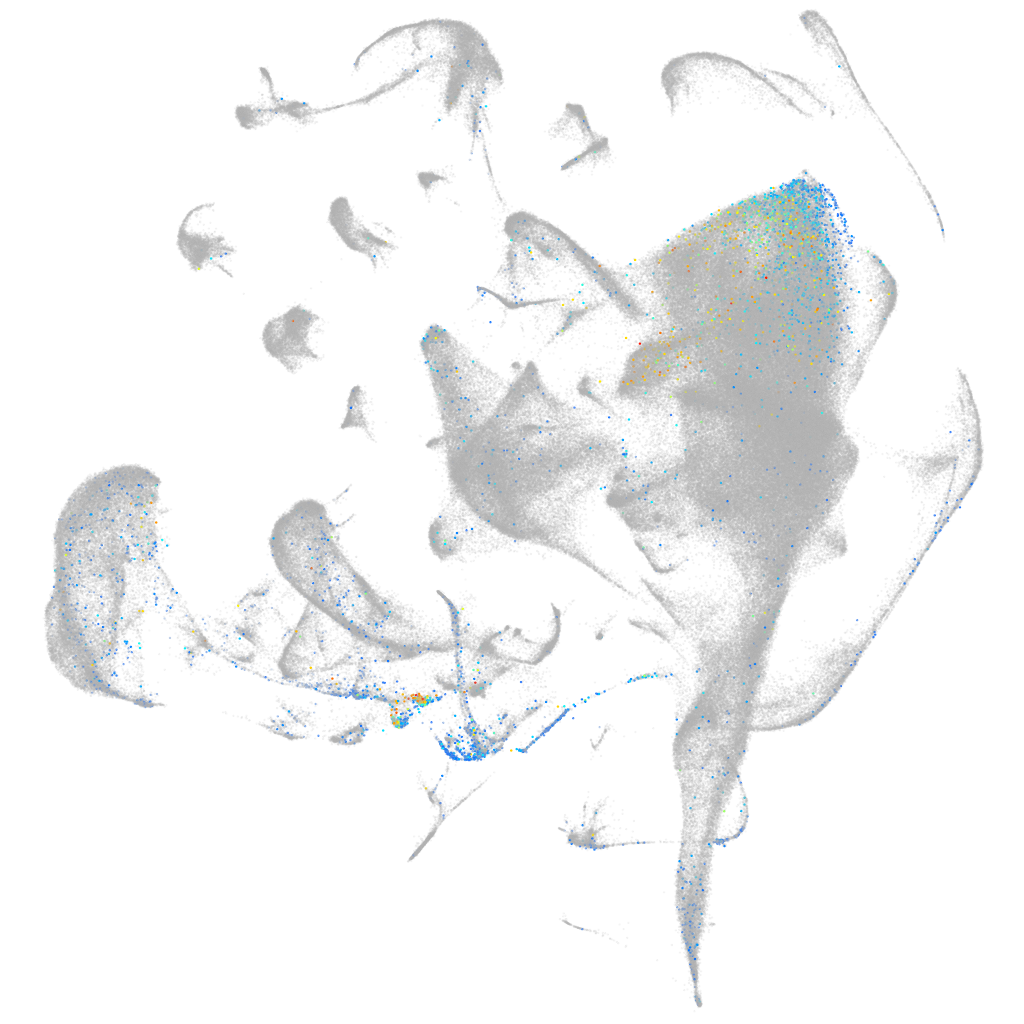synaptotagmin-like 5
ZFIN 
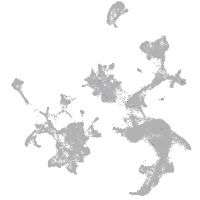
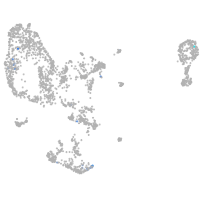
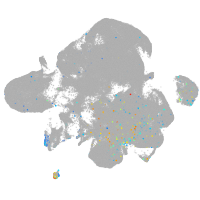
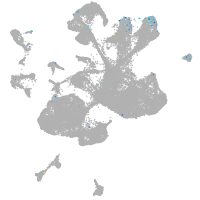
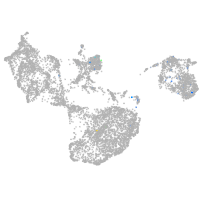
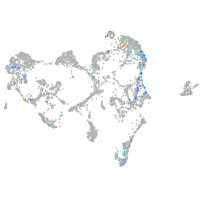
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cldnh | 0.114 | msna | -0.034 |
| gfral | 0.110 | akap12b | -0.030 |
| vill | 0.107 | mdka | -0.030 |
| vtcn1 | 0.099 | cx43.4 | -0.029 |
| itpr3 | 0.098 | hspb1 | -0.029 |
| CR759836.1 | 0.093 | ccnb1 | -0.025 |
| prr15la | 0.093 | stm | -0.025 |
| fer1l4 | 0.091 | apoc1 | -0.024 |
| calm1b | 0.089 | id1 | -0.024 |
| grp | 0.088 | lig1 | -0.024 |
| lad1 | 0.084 | mki67 | -0.024 |
| si:ch211-256e16.6 | 0.084 | sb:cb81 | -0.024 |
| CABZ01073083.1 | 0.083 | pou5f3 | -0.023 |
| foxa3 | 0.083 | sept12 | -0.023 |
| pou2f3 | 0.083 | si:ch211-152c2.3 | -0.023 |
| zgc:92380 | 0.083 | smo | -0.023 |
| stxbp1a | 0.081 | sox19a | -0.023 |
| snap25a | 0.080 | tpx2 | -0.023 |
| ywhag2 | 0.080 | vox | -0.023 |
| agr2 | 0.079 | XLOC-003690 | -0.023 |
| hepacam2 | 0.079 | ccna2 | -0.022 |
| cnfn | 0.078 | chaf1a | -0.022 |
| eno2 | 0.078 | gpm6bb | -0.022 |
| st6galnac1.1 | 0.078 | lbr | -0.022 |
| sv2a | 0.078 | lfng | -0.022 |
| cldn8 | 0.077 | notch3 | -0.022 |
| scg2a | 0.077 | tspan7 | -0.022 |
| stx1b | 0.077 | ved | -0.022 |
| vamp2 | 0.077 | anp32e | -0.021 |
| mir375-2 | 0.076 | cdca8 | -0.021 |
| si:ch211-130m23.2 | 0.075 | dnmt3bb.2 | -0.021 |
| syn2a | 0.074 | fzd7a | -0.021 |
| cplx2 | 0.073 | hbbe2 | -0.021 |
| rab6bb | 0.073 | nop58 | -0.021 |
| sytl4 | 0.073 | notch1a | -0.021 |