suppressor of variegation 3-9 homolog 1b
ZFIN 
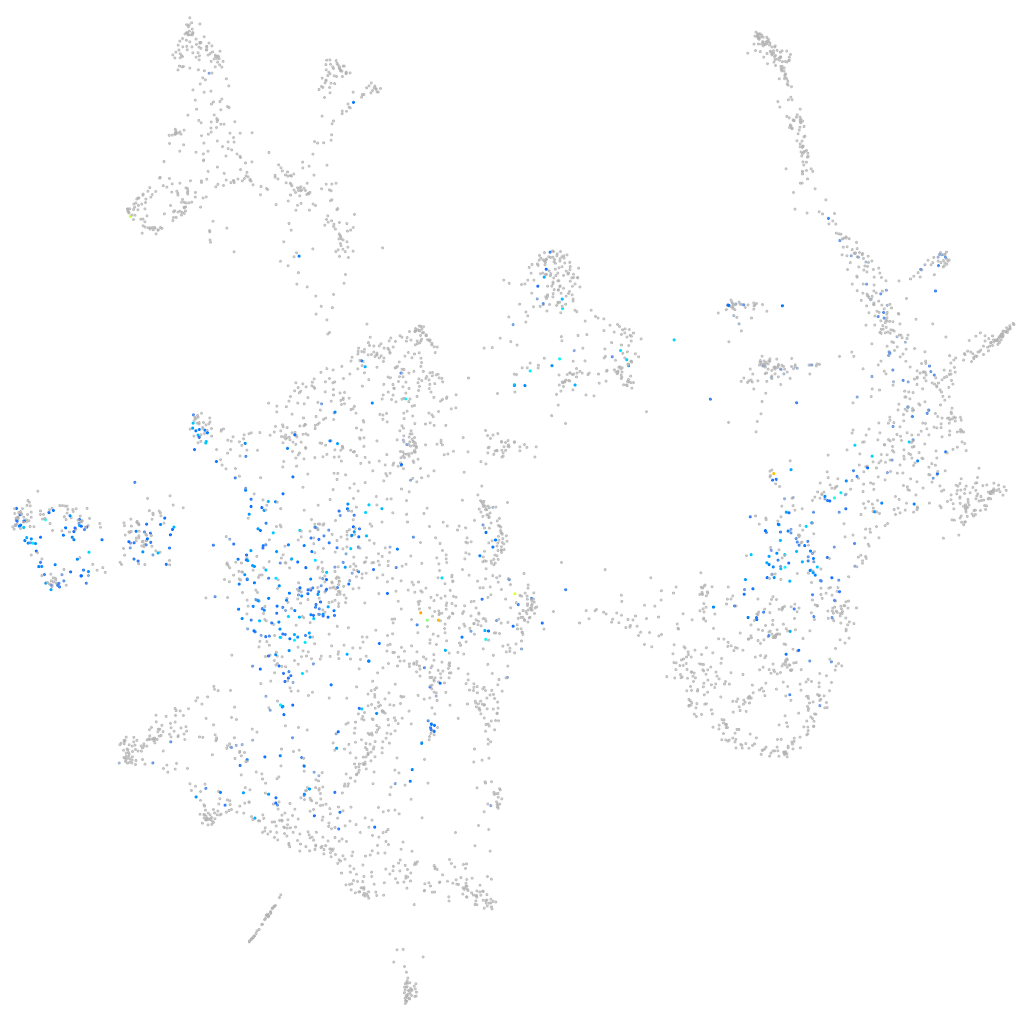
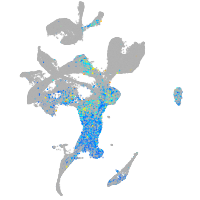
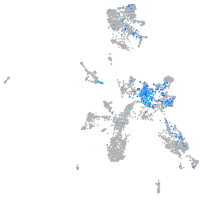
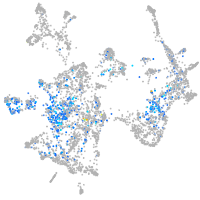
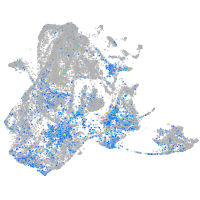
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pcna | 0.501 | tob1b | -0.160 |
| rrm2 | 0.489 | h1f0 | -0.157 |
| dut | 0.485 | b2ml | -0.145 |
| rpa2 | 0.478 | nupr1a | -0.142 |
| zgc:110540 | 0.455 | calm1a | -0.141 |
| rrm1 | 0.454 | tmem59 | -0.140 |
| slbp | 0.450 | atp2b1a | -0.140 |
| fen1 | 0.437 | eno1a | -0.134 |
| nasp | 0.435 | spint2 | -0.134 |
| stmn1a | 0.433 | CR383676.1 | -0.132 |
| CABZ01005379.1 | 0.426 | mt-co2 | -0.130 |
| chaf1a | 0.423 | otofb | -0.129 |
| dnajc9 | 0.414 | atp6v1e1b | -0.128 |
| cks1b | 0.411 | calml4a | -0.126 |
| mibp | 0.410 | pkig | -0.125 |
| rpa3 | 0.410 | ccni | -0.125 |
| dhfr | 0.410 | atp1a3b | -0.124 |
| unga | 0.409 | cd164l2 | -0.124 |
| asf1ba | 0.409 | gapdhs | -0.124 |
| prim2 | 0.397 | dnajc5b | -0.124 |
| rfc4 | 0.391 | pvalb8 | -0.124 |
| lig1 | 0.390 | rnasekb | -0.123 |
| tubb2b | 0.390 | nptna | -0.123 |
| rbbp4 | 0.387 | prr15la | -0.122 |
| dek | 0.384 | gabarapl2 | -0.121 |
| sumo3b | 0.382 | calm1b | -0.120 |
| esco2 | 0.379 | igfbp5b | -0.120 |
| rnaseh2a | 0.378 | gpx2 | -0.120 |
| orc4 | 0.375 | CR925719.1 | -0.119 |
| shmt1 | 0.375 | capgb | -0.118 |
| LOC100330864 | 0.373 | osbpl1a | -0.118 |
| ccna2 | 0.372 | COX3 | -0.117 |
| anp32b | 0.371 | adipor2 | -0.117 |
| zgc:110216 | 0.371 | tspan13b | -0.116 |
| si:dkey-6i22.5 | 0.368 | twf2b | -0.116 |