suppressor of variegation 3-9 homolog 1b
ZFIN 
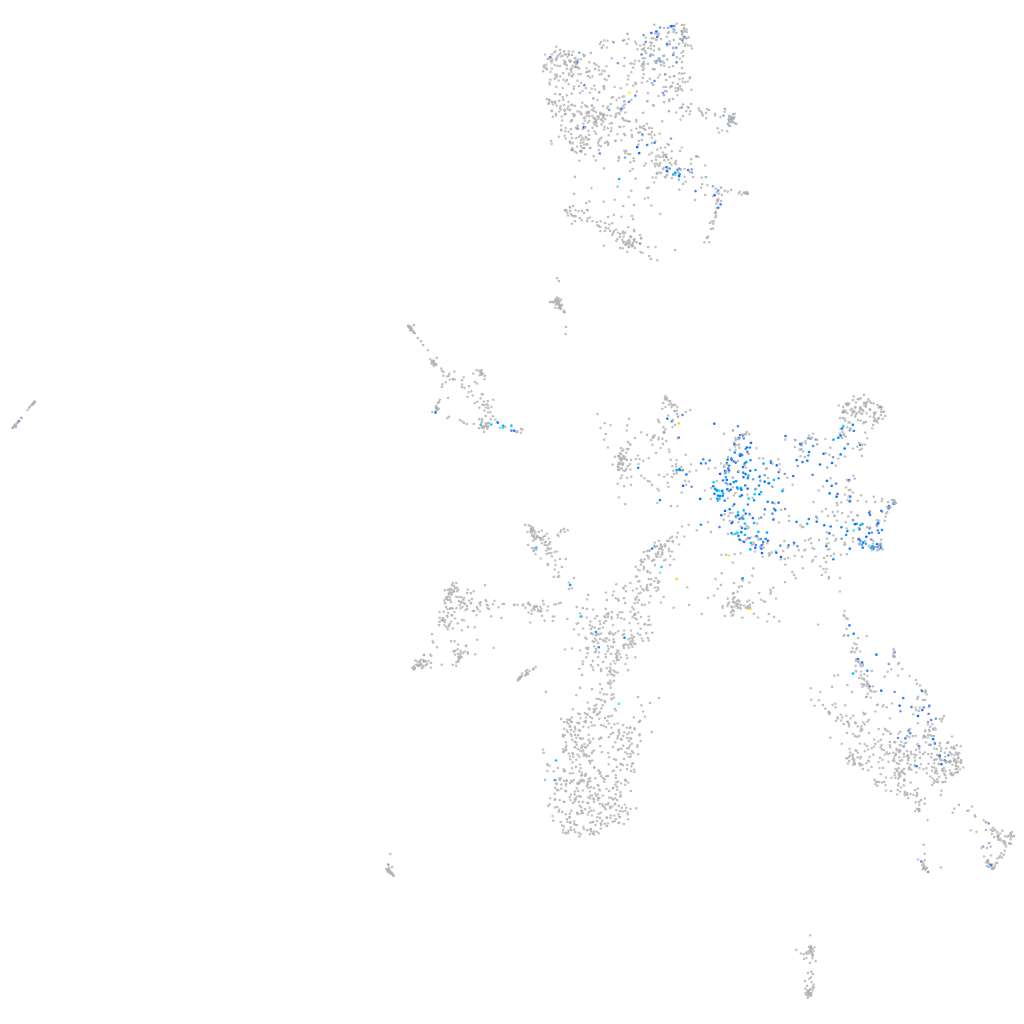
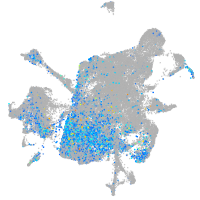
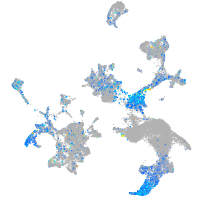
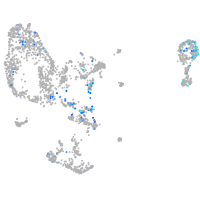
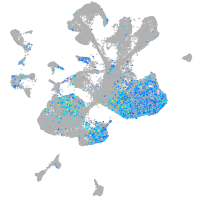
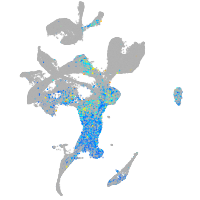
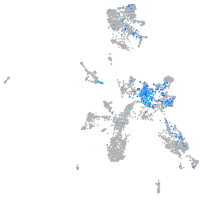
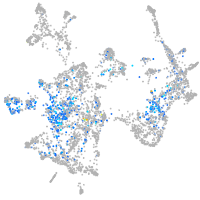
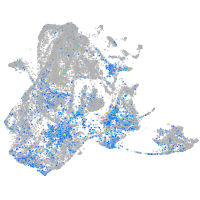
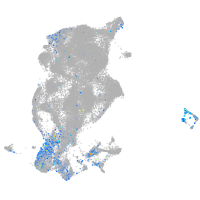
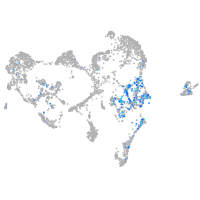
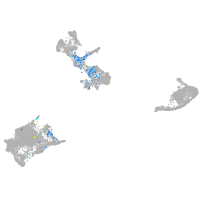
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:110540 | 0.515 | txn | -0.274 |
| rrm2 | 0.496 | gstp1 | -0.251 |
| dut | 0.495 | anxa5b | -0.235 |
| rrm1 | 0.485 | cldnh | -0.226 |
| CABZ01005379.1 | 0.485 | gapdhs | -0.225 |
| mibp | 0.479 | CR383676.1 | -0.215 |
| rpa2 | 0.471 | calm1a | -0.215 |
| stmn1a | 0.466 | tmem59 | -0.213 |
| nasp | 0.461 | wu:fb18f06 | -0.210 |
| pcna | 0.458 | gabarapa | -0.204 |
| chaf1a | 0.454 | selenow2b | -0.202 |
| ccna2 | 0.443 | fxyd1 | -0.200 |
| cks1b | 0.442 | vamp2 | -0.197 |
| slbp | 0.439 | rnasekb | -0.197 |
| mcm7 | 0.438 | atp6v1e1b | -0.197 |
| si:dkey-6i22.5 | 0.437 | selenow1 | -0.195 |
| lbr | 0.437 | rtn1a | -0.187 |
| zgc:110216 | 0.431 | pvalb5 | -0.187 |
| mis12 | 0.424 | bik | -0.186 |
| cdca5 | 0.415 | nfe2l2a | -0.185 |
| fbxo5 | 0.414 | chga | -0.184 |
| esco2 | 0.412 | smdt1b | -0.181 |
| shmt1 | 0.411 | itm2ba | -0.181 |
| ncapg | 0.409 | si:ch211-153b23.5 | -0.178 |
| rpa3 | 0.408 | atp6v0cb | -0.178 |
| prim2 | 0.406 | gnb1a | -0.176 |
| lig1 | 0.405 | camk2n1a | -0.172 |
| haus4 | 0.404 | icn | -0.172 |
| atad5a | 0.403 | ip6k2a | -0.170 |
| smc2 | 0.402 | htatip2 | -0.169 |
| tk1 | 0.400 | olfm1b | -0.169 |
| dhfr | 0.398 | COX3 | -0.169 |
| mki67 | 0.396 | si:dkeyp-75h12.5 | -0.168 |
| asf1ba | 0.393 | si:ch1073-443f11.2 | -0.167 |
| si:dkey-261m9.17 | 0.392 | scg2b | -0.167 |