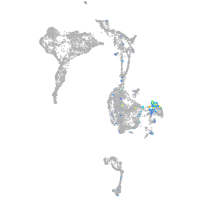
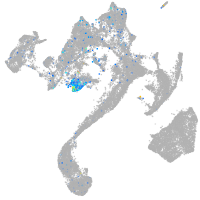
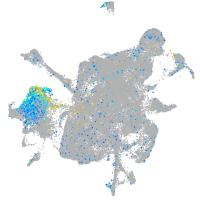
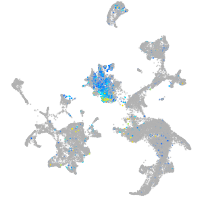
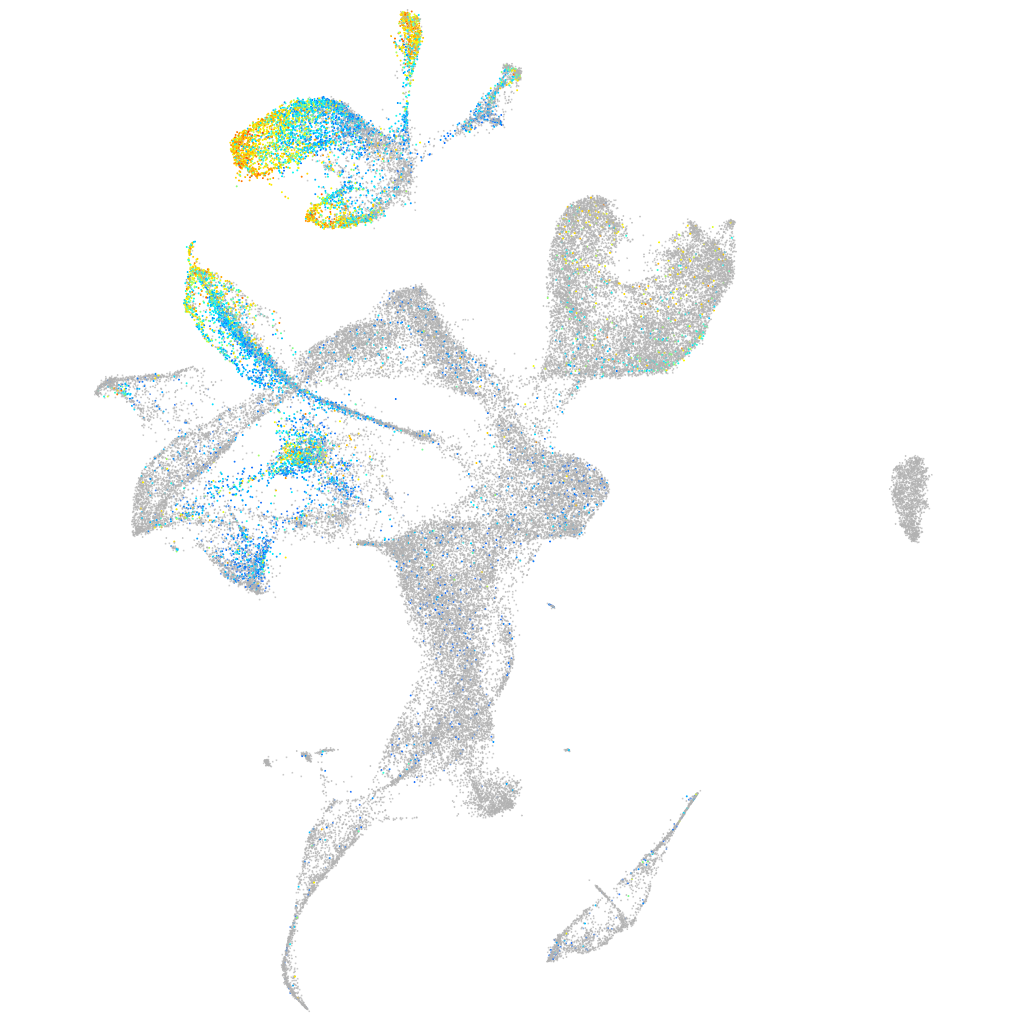syntaxin binding protein 1a
ZFIN 
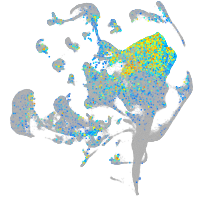
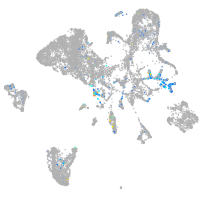
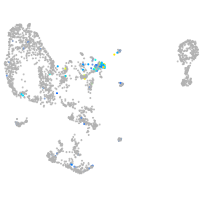
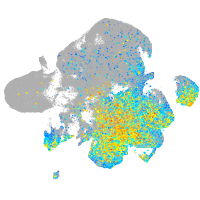
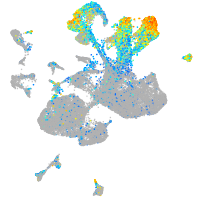
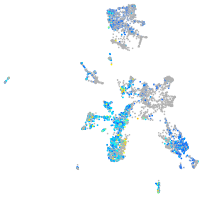
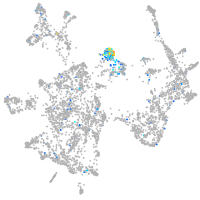
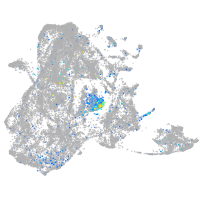
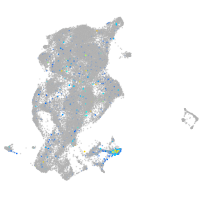
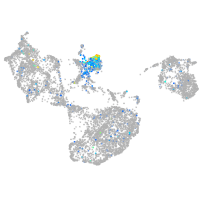
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| syt1a | 0.612 | hmgb2a | -0.417 |
| stx1b | 0.584 | hmga1a | -0.234 |
| ywhag2 | 0.568 | ahcy | -0.227 |
| slc32a1 | 0.546 | pcna | -0.224 |
| sv2a | 0.543 | stmn1a | -0.220 |
| eno2 | 0.535 | mdka | -0.218 |
| sncb | 0.516 | msi1 | -0.213 |
| elavl3 | 0.515 | msna | -0.209 |
| rbfox1 | 0.501 | rrm1 | -0.207 |
| slc6a1a | 0.499 | hmgb2b | -0.205 |
| mir181b-3 | 0.494 | nutf2l | -0.205 |
| si:ch73-119p20.1 | 0.494 | dut | -0.203 |
| stmn1b | 0.487 | fabp7a | -0.202 |
| gng3 | 0.485 | mki67 | -0.200 |
| rtn1b | 0.476 | lbr | -0.199 |
| gabrb4 | 0.472 | mcm7 | -0.198 |
| snap25a | 0.456 | ccnd1 | -0.197 |
| zgc:65894 | 0.449 | chaf1a | -0.197 |
| sprn | 0.433 | tuba8l4 | -0.196 |
| tkta | 0.431 | tuba8l | -0.194 |
| syt2a | 0.430 | selenoh | -0.189 |
| zgc:153426 | 0.430 | ccna2 | -0.188 |
| nrxn1a | 0.426 | her15.1 | -0.186 |
| syn2a | 0.425 | banf1 | -0.185 |
| slc35g2b | 0.420 | dek | -0.184 |
| tfap2a | 0.419 | rps20 | -0.184 |
| slc6a1b | 0.417 | eef1da | -0.181 |
| st8sia5 | 0.415 | rpsa | -0.179 |
| si:ch211-129p13.1 | 0.413 | cks1b | -0.178 |
| vsnl1b | 0.413 | rplp2l | -0.177 |
| vamp2 | 0.412 | rpa3 | -0.177 |
| celf5a | 0.411 | rpl12 | -0.177 |
| LOC101886114 | 0.410 | rplp1 | -0.175 |
| gnao1a | 0.407 | lig1 | -0.175 |
| stmn2a | 0.405 | cx43.4 | -0.175 |