sorcin
ZFIN 
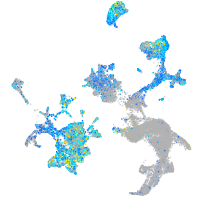
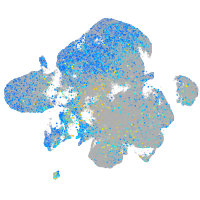
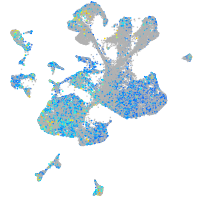
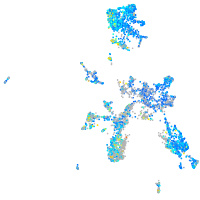
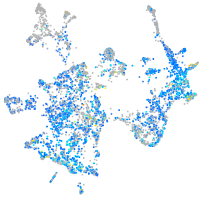
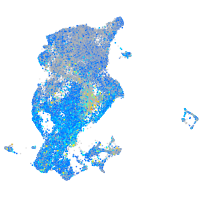
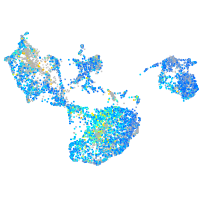
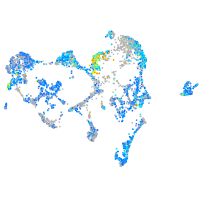
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| bsnd | 0.417 | hspb1 | -0.198 |
| malb | 0.389 | wu:fb97g03 | -0.194 |
| phlda2 | 0.386 | pabpc1a | -0.187 |
| si:ch73-359m17.9 | 0.379 | eif5a2 | -0.181 |
| gdpd3a | 0.379 | marcksb | -0.176 |
| clcnk | 0.378 | fbl | -0.174 |
| mal2 | 0.371 | cx43.4 | -0.171 |
| agr2 | 0.364 | apoc1 | -0.170 |
| zgc:158423 | 0.362 | apoeb | -0.170 |
| si:dkey-36i7.3 | 0.352 | hnrnpa1b | -0.168 |
| trim35-12 | 0.351 | hnrnpabb | -0.162 |
| rnf183 | 0.350 | snu13b | -0.162 |
| atp1a1a.3 | 0.349 | ncl | -0.158 |
| bin2a | 0.349 | cth | -0.157 |
| tmem72 | 0.347 | pdzk1ip1 | -0.156 |
| sb:cb1058 | 0.345 | top1l | -0.152 |
| cldnh | 0.345 | gpt2l | -0.152 |
| ccl19a.1 | 0.340 | zmp:0000000624 | -0.151 |
| cd63 | 0.337 | nono | -0.149 |
| soul2 | 0.330 | ilf3b | -0.149 |
| ndufa4 | 0.326 | hnrnpa1a | -0.149 |
| ctgfa | 0.323 | lbx2 | -0.149 |
| ppp1r1b | 0.322 | nhp2 | -0.148 |
| gucy2c | 0.318 | parp1 | -0.148 |
| elf3 | 0.315 | cdx4 | -0.146 |
| ahnak | 0.313 | hmgb2b | -0.145 |
| AL954322.2 | 0.312 | slc5a12 | -0.141 |
| tbx2b | 0.312 | agxtb | -0.141 |
| vdac2 | 0.311 | ppiaa | -0.140 |
| etf1a | 0.311 | znfl2a | -0.140 |
| me3 | 0.310 | brd3a | -0.140 |
| sfxn3 | 0.308 | NC-002333.4 | -0.140 |
| krt8 | 0.307 | si:dkey-194e6.1 | -0.139 |
| cox6b1 | 0.307 | marcksl1b | -0.139 |
| oclnb | 0.303 | ASS1 | -0.139 |