sperm associated antigen 9a
ZFIN 
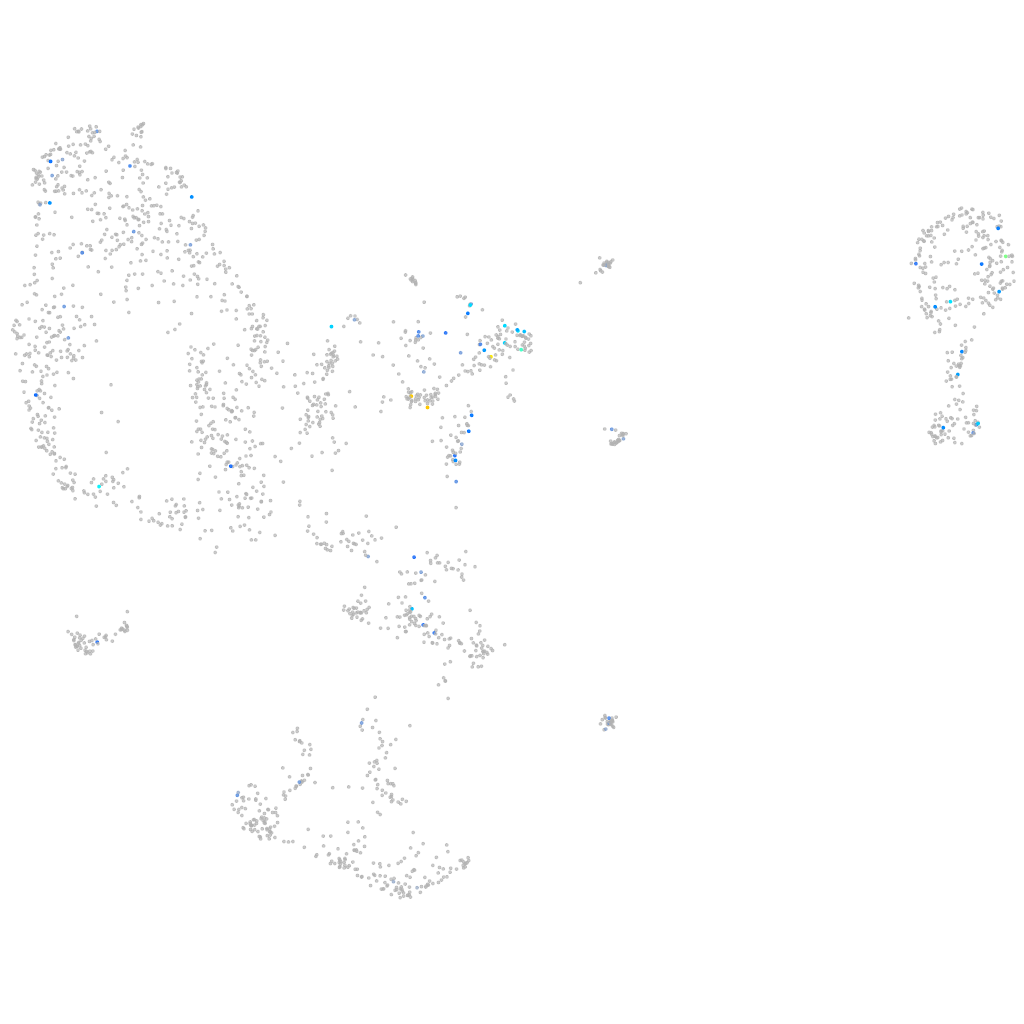
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| XLOC-025875 | 0.296 | cox5aa | -0.114 |
| CABZ01044023.1 | 0.295 | cox7b | -0.112 |
| trim101 | 0.285 | cox6a1 | -0.110 |
| tmem266 | 0.274 | atp5f1b | -0.109 |
| camta1 | 0.270 | rpl35a | -0.103 |
| CR759927.2 | 0.265 | atp5fa1 | -0.101 |
| taok3b | 0.247 | ndufb8 | -0.100 |
| prdm13 | 0.244 | uqcrh | -0.097 |
| LOC101884310 | 0.242 | romo1 | -0.094 |
| ripk2 | 0.241 | GCA | -0.094 |
| slc7a14a | 0.236 | epcam | -0.093 |
| osbp2 | 0.236 | ndufa12 | -0.093 |
| pak7 | 0.236 | slc25a5 | -0.091 |
| erbb4a | 0.233 | ndrg1a | -0.091 |
| ndrg4 | 0.231 | sypl2a | -0.091 |
| atp6v0cb | 0.228 | glod5 | -0.091 |
| her13 | 0.227 | cyc1 | -0.090 |
| stxbp5a | 0.218 | cox6b2 | -0.089 |
| zmp:0000000760 | 0.216 | glrx | -0.089 |
| fryb | 0.215 | ndufs6 | -0.089 |
| map2 | 0.214 | atp1b1a | -0.088 |
| sox12 | 0.212 | ndufa5 | -0.087 |
| kcnc3b | 0.211 | cdaa | -0.087 |
| mapk11 | 0.210 | mt-nd2 | -0.087 |
| pitx2 | 0.210 | ndufc2 | -0.086 |
| dhx32b | 0.209 | mrps36 | -0.086 |
| nova2 | 0.208 | vamp3 | -0.085 |
| elmo2 | 0.208 | phb2b | -0.085 |
| hecw1b | 0.208 | cox4i1 | -0.085 |
| CABZ01041604.1 | 0.207 | hadhb | -0.085 |
| pitx3 | 0.207 | sgk2a | -0.083 |
| rab25a | 0.203 | cd9b | -0.083 |
| CR974456.1 | 0.202 | krt18a.1 | -0.082 |
| SLCO3A1 (1 of many) | 0.201 | myl12.1 | -0.080 |
| vamp2 | 0.200 | si:rp71-17i16.6 | -0.080 |