sorting nexin 19a
ZFIN 
Other cell groups


all cells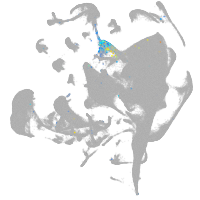 |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 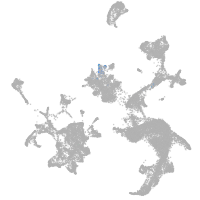 |
pronephros 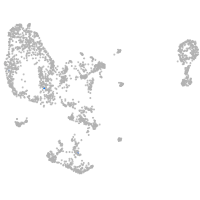 |
neural |
spinal cord / glia |
eye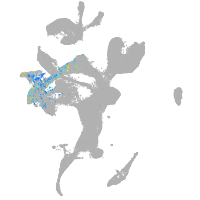 |
taste / olfactory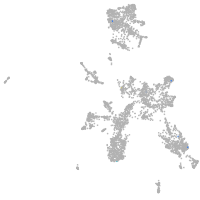 |
otic / lateral line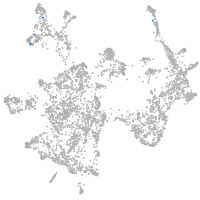 |
epidermis |
periderm |
fin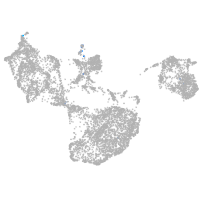 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tmx3a | 0.289 | hbbe3 | -0.021 |
| si:ch1073-469d17.2 | 0.284 | rab5c | -0.018 |
| slc25a3a | 0.281 | arl4aa | -0.018 |
| arl3l2 | 0.275 | lmo2 | -0.017 |
| ckmt2a | 0.271 | arhgdia | -0.016 |
| glmnb | 0.267 | yrk | -0.016 |
| rbp4l | 0.262 | tuba8l4 | -0.016 |
| zgc:73359 | 0.261 | etv2 | -0.016 |
| si:ch211-175f12.2 | 0.259 | capgb | -0.016 |
| rcvrna | 0.257 | pycard | -0.015 |
| prph2a | 0.255 | atp1b1a | -0.015 |
| grk1b | 0.253 | rhocb | -0.015 |
| guk1b | 0.245 | myct1a | -0.015 |
| gnat2 | 0.245 | zgc:162730 | -0.014 |
| si:dkey-17e16.15 | 0.244 | junba | -0.014 |
| elovl4b | 0.244 | mob1a | -0.014 |
| rcvrn2 | 0.243 | arhgdig | -0.014 |
| arr3a | 0.241 | csrp1a | -0.014 |
| pde6c | 0.239 | srgn | -0.014 |
| rcvrn3 | 0.239 | slc25a3b | -0.014 |
| cngb3.2 | 0.234 | reep5 | -0.014 |
| si:ch211-103a14.5 | 0.231 | rnaseka | -0.014 |
| si:ch211-81a5.8 | 0.231 | mafbb | -0.014 |
| cnga3a | 0.231 | si:ch211-114n24.6 | -0.013 |
| gnb3b | 0.225 | zgc:158343 | -0.013 |
| arl3l1 | 0.224 | alad | -0.013 |
| tmem237a | 0.218 | ehd4 | -0.013 |
| mapkapk2b | 0.217 | ckma | -0.013 |
| guca1c | 0.217 | si:ch73-248e21.7 | -0.013 |
| pdcb | 0.214 | cnn2 | -0.013 |
| zgc:103625 | 0.211 | fam53b | -0.013 |
| opn6a | 0.207 | ctsz | -0.013 |
| syt5a | 0.204 | abracl | -0.013 |
| si:dkey-94e7.2 | 0.203 | si:ch1073-392o20.2 | -0.013 |
| grk7a | 0.200 | scarf1 | -0.013 |




