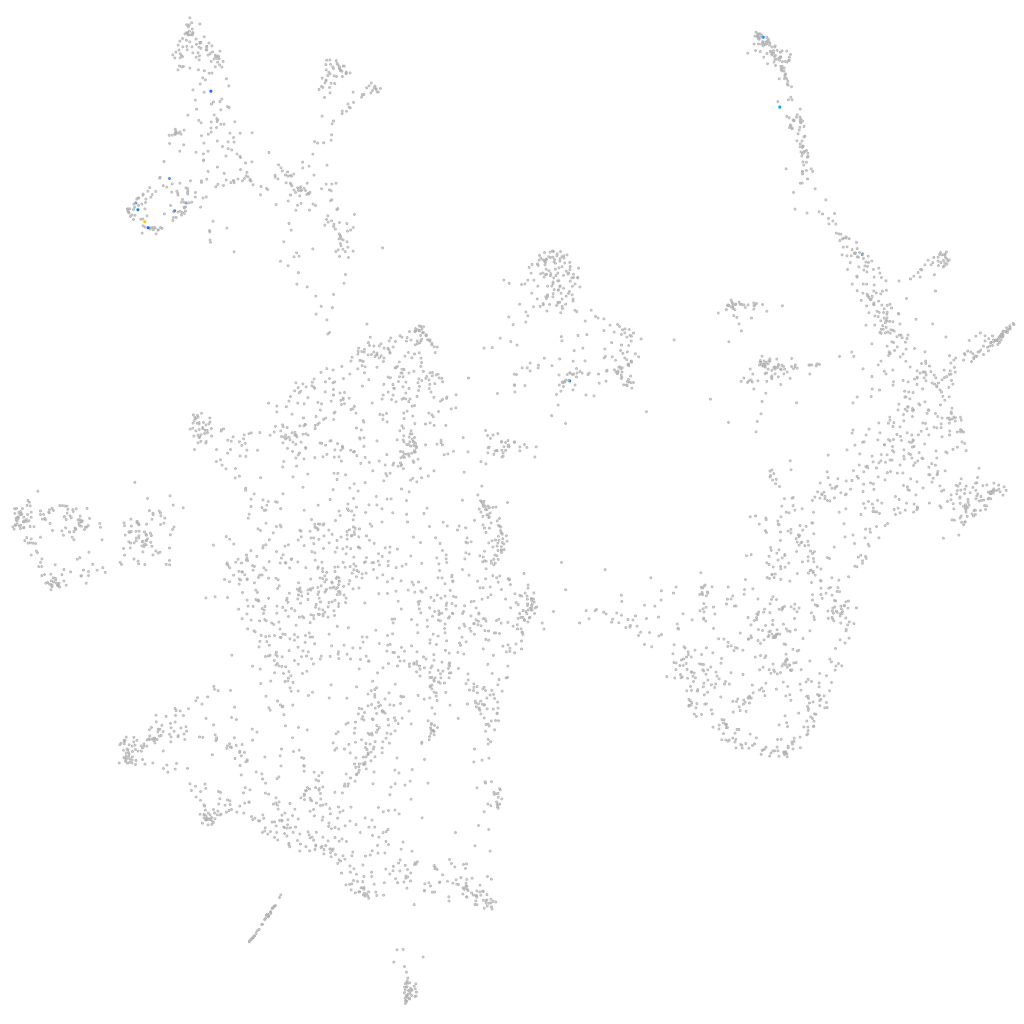
sorting nexin 19a
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye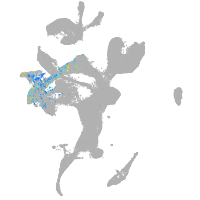 |
taste / olfactory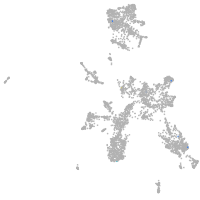 |
otic / lateral line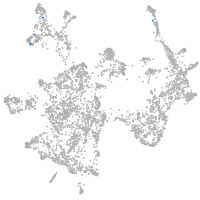 |
epidermis |
periderm |
fin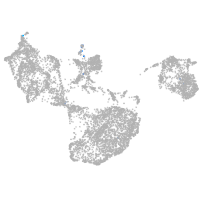 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC108190653 | 0.681 | rps19 | -0.115 |
| gzmk | 0.462 | rplp1 | -0.115 |
| AL953886.2 | 0.428 | rpl28 | -0.112 |
| CABZ01024499.1 | 0.376 | rps14 | -0.109 |
| LOC100001665 | 0.367 | rpl26 | -0.105 |
| dicp1.5-6 | 0.313 | rps24 | -0.102 |
| cep126 | 0.303 | rpl39 | -0.101 |
| p2ry12 | 0.298 | rpl14 | -0.100 |
| slc24a5 | 0.294 | rps15a | -0.099 |
| LOC110438243 | 0.294 | si:dkey-151g10.6 | -0.098 |
| EML6 | 0.274 | rps23 | -0.098 |
| hnf4b | 0.271 | rpl12 | -0.097 |
| CABZ01083442.1 | 0.264 | rpl35a | -0.097 |
| RF01268 | 0.256 | rplp2l | -0.097 |
| SYNDIG1 | 0.252 | nme2b.1 | -0.096 |
| or133-9 | 0.247 | rpl13 | -0.096 |
| nfat5b | 0.246 | rps15 | -0.093 |
| march1 | 0.235 | rpl10a | -0.092 |
| SAT2 | 0.234 | rpl11 | -0.092 |
| CU633486.1 | 0.233 | rps26l | -0.091 |
| tceb2 | 0.232 | rps21 | -0.091 |
| ATE1 | 0.227 | rpl18 | -0.090 |
| dars2 | 0.227 | rpl7 | -0.090 |
| acot7 | 0.222 | rpl10 | -0.089 |
| dnai2b | 0.221 | rps26 | -0.087 |
| CABZ01079663.1 | 0.221 | rps3a | -0.087 |
| LOC101882062 | 0.219 | rpl7a | -0.087 |
| rnf139 | 0.218 | rps8a | -0.086 |
| arhgap23b | 0.218 | rpl17 | -0.086 |
| tns1a | 0.217 | rpl9 | -0.086 |
| pfkfb2b | 0.216 | rps20 | -0.085 |
| cntnap2a | 0.216 | rpl32 | -0.084 |
| bbox1 | 0.215 | rpl24 | -0.082 |
| ccser2b | 0.214 | rps28 | -0.082 |
| plppr4b | 0.206 | naca | -0.080 |