snail family zinc finger 1b
ZFIN 
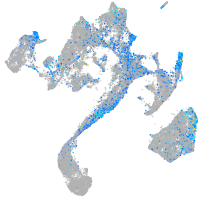
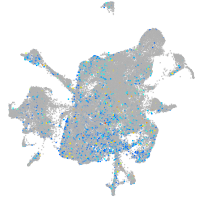
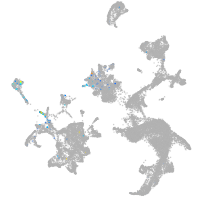
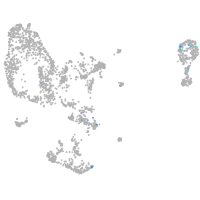
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ednrab | 0.665 | nova2 | -0.095 |
| tfec | 0.504 | tuba1c | -0.091 |
| BX248318.1 | 0.392 | gpm6aa | -0.086 |
| si:ch211-11c3.9 | 0.344 | ckbb | -0.071 |
| crestin | 0.339 | rtn1a | -0.070 |
| si:ch211-11c3.12 | 0.292 | ptmaa | -0.065 |
| msx2b | 0.290 | sox3 | -0.065 |
| crlf1a | 0.275 | elavl3 | -0.064 |
| sfrp2 | 0.258 | CU467822.1 | -0.059 |
| foxd3 | 0.248 | fabp7a | -0.059 |
| itga9 | 0.244 | vim | -0.059 |
| inka1a | 0.231 | tmsb | -0.059 |
| XLOC-042222 | 0.229 | gapdhs | -0.058 |
| wu:fb97g03 | 0.219 | CR383676.1 | -0.058 |
| ets1 | 0.214 | mdka | -0.057 |
| msx1b | 0.214 | rnasekb | -0.056 |
| dlc1 | 0.213 | gfap | -0.055 |
| tfap2a | 0.213 | zgc:165461 | -0.054 |
| si:ch211-107n13.1 | 0.210 | cadm3 | -0.053 |
| nid2a | 0.204 | sox19a | -0.053 |
| erbb3b | 0.204 | si:dkey-56m19.5 | -0.053 |
| kcnk5b | 0.202 | pvalb1 | -0.053 |
| XLOC-001603 | 0.200 | ncam1a | -0.052 |
| pdgfra | 0.196 | CU634008.1 | -0.051 |
| LOC110439533 | 0.193 | si:ch211-137a8.4 | -0.050 |
| slc15a2 | 0.191 | tox | -0.050 |
| nid1b | 0.188 | stmn1b | -0.049 |
| BX927258.1 | 0.179 | fam168a | -0.048 |
| LOC110438542 | 0.172 | actc1b | -0.048 |
| hmcn1 | 0.170 | tspan3a | -0.048 |
| nradd | 0.166 | nr2f1a | -0.048 |
| cyth1b | 0.161 | pvalb2 | -0.048 |
| zgc:165582 | 0.153 | hmgb1b | -0.048 |
| cysltr1 | 0.149 | slc1a2b | -0.047 |
| si:dkeyp-59c12.1 | 0.148 | myt1b | -0.047 |