small integral membrane protein 29
ZFIN 
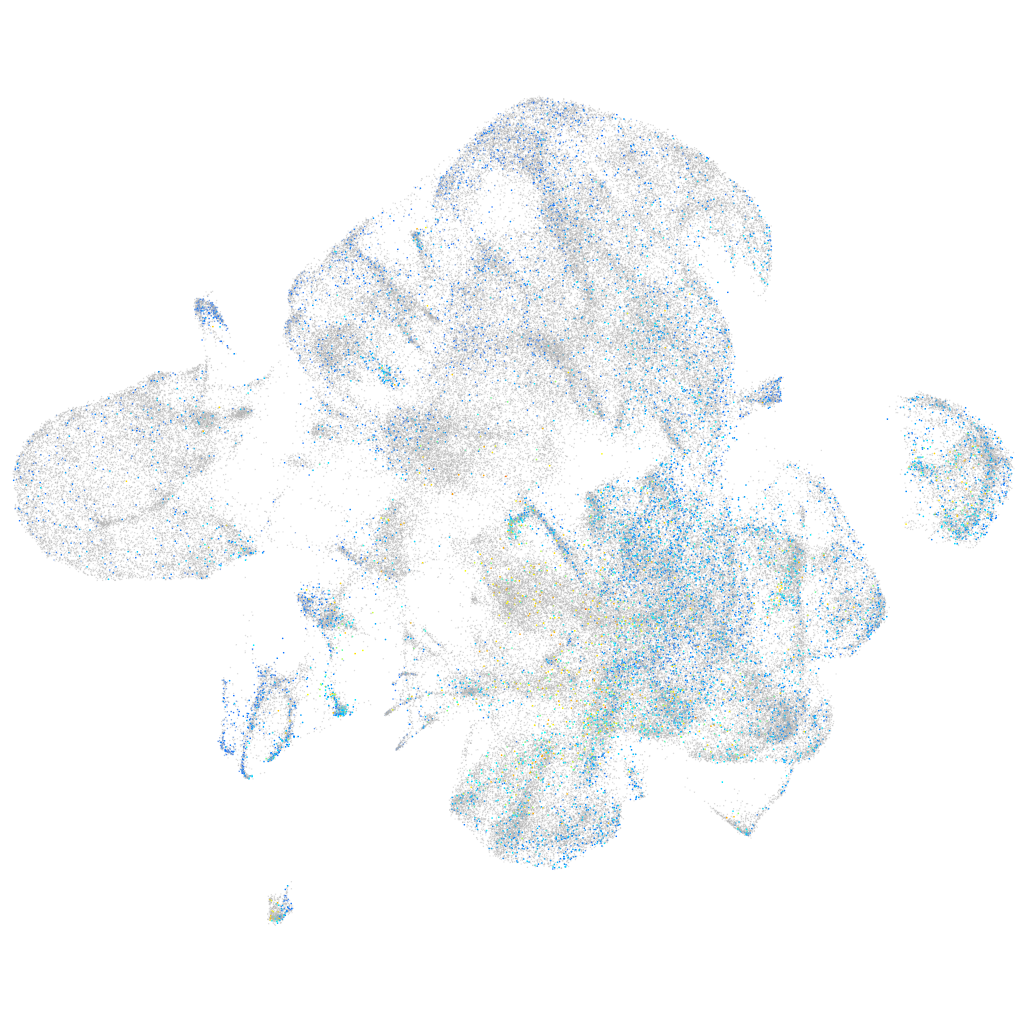
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| atp6v1e1b | 0.108 | hmgb2a | -0.107 |
| atp6v1g1 | 0.104 | pcna | -0.088 |
| calm1a | 0.104 | stmn1a | -0.088 |
| ppdpfb | 0.103 | hmgb2b | -0.080 |
| gng3 | 0.103 | anp32b | -0.077 |
| stxbp1a | 0.102 | banf1 | -0.077 |
| vamp2 | 0.101 | mki67 | -0.077 |
| rnasekb | 0.100 | id1 | -0.076 |
| sncb | 0.099 | tuba8l4 | -0.075 |
| snap25a | 0.097 | chaf1a | -0.075 |
| gabarapb | 0.096 | lig1 | -0.074 |
| rtn1a | 0.096 | si:ch73-281n10.2 | -0.073 |
| mllt11 | 0.096 | ranbp1 | -0.072 |
| stmn1b | 0.095 | msna | -0.072 |
| gabarapl2 | 0.095 | nop58 | -0.072 |
| atp6v0cb | 0.094 | tuba8l | -0.072 |
| gapdhs | 0.090 | nasp | -0.071 |
| necap1 | 0.090 | hmga1a | -0.070 |
| ywhah | 0.089 | rrm1 | -0.070 |
| gnb1a | 0.089 | lbr | -0.070 |
| ywhag2 | 0.088 | ccnd1 | -0.069 |
| csdc2a | 0.088 | dkc1 | -0.069 |
| gng2 | 0.088 | ccna2 | -0.068 |
| hist2h2l | 0.088 | mcm7 | -0.067 |
| gabarapa | 0.087 | rpa3 | -0.067 |
| vdac3 | 0.087 | nop56 | -0.066 |
| h2afx1 | 0.087 | fen1 | -0.066 |
| calm2a | 0.087 | dek | -0.066 |
| zgc:65894 | 0.086 | snu13b | -0.066 |
| stmn2a | 0.086 | sox19a | -0.065 |
| stx1b | 0.086 | dut | -0.065 |
| sh3gl2a | 0.086 | npm1a | -0.065 |
| si:dkeyp-75h12.5 | 0.085 | fbl | -0.064 |
| tpi1b | 0.085 | ssrp1a | -0.064 |
| eno2 | 0.084 | mcm6 | -0.064 |