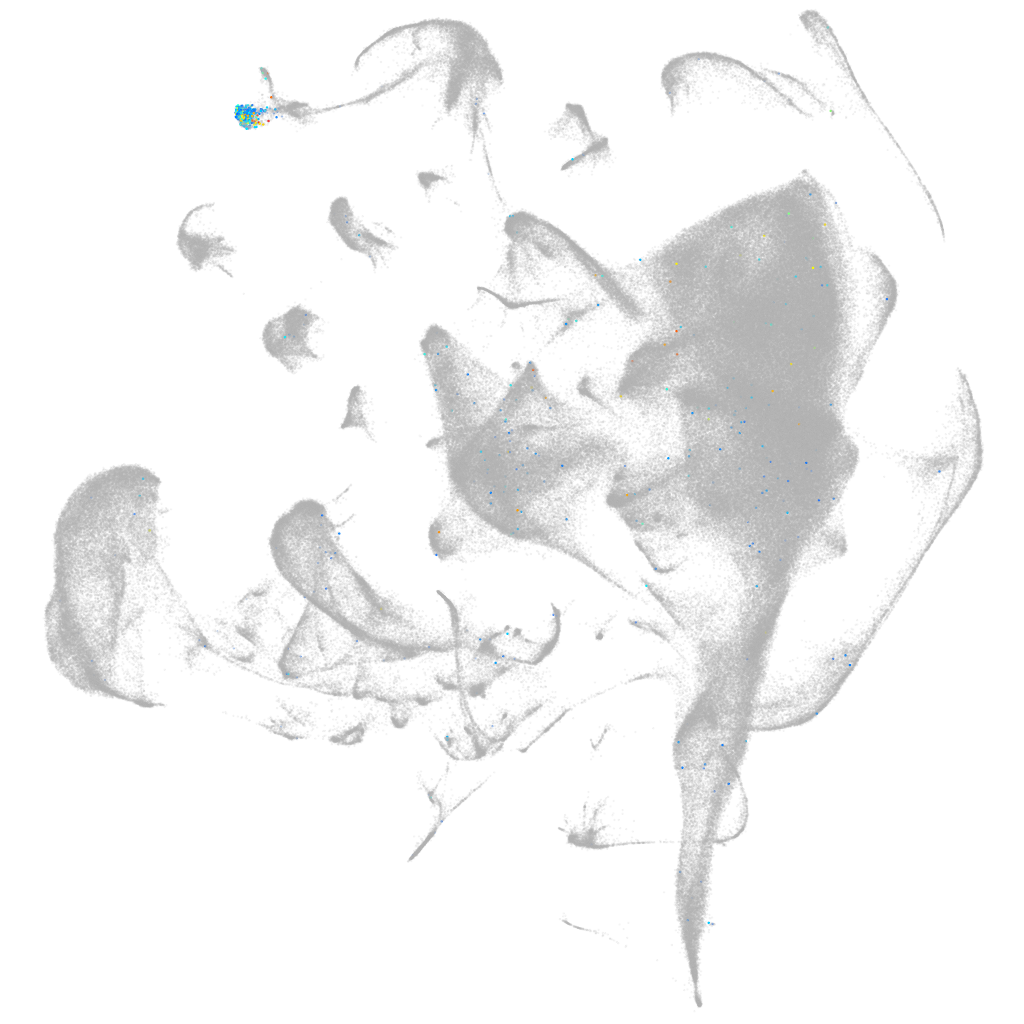
solute carrier family 2 member 6
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cx32.2 | 0.530 | fabp3 | -0.036 |
| mpeg1.1 | 0.504 | hmgb1b | -0.035 |
| si:ch211-147m6.1 | 0.500 | hmgb3a | -0.030 |
| si:ch211-194m7.3 | 0.499 | jpt1b | -0.029 |
| havcr1 | 0.486 | mdka | -0.028 |
| ccl34a.4 | 0.483 | pfn2l | -0.028 |
| cmklr1 | 0.483 | nova2 | -0.026 |
| mfap4 | 0.466 | tuba1c | -0.026 |
| si:dkey-148a17.6 | 0.463 | vdac1 | -0.026 |
| LOC108180725 | 0.457 | marcksb | -0.025 |
| si:dkey-5n18.1 | 0.452 | tuba1a | -0.024 |
| spi1a | 0.441 | si:ch211-137a8.4 | -0.023 |
| marco | 0.434 | tmeff1b | -0.023 |
| rnaset2l | 0.426 | apex1 | -0.022 |
| c1qb | 0.419 | gpm6aa | -0.022 |
| cxcr3.2 | 0.415 | hnrnpa0a | -0.022 |
| itgae.1 | 0.408 | mdkb | -0.022 |
| grna | 0.398 | tspan7 | -0.022 |
| zgc:173915 | 0.395 | foxp4 | -0.021 |
| fcer1gl | 0.392 | h2afva | -0.021 |
| si:ch211-1o7.3 | 0.390 | meis1b | -0.021 |
| adgrg3 | 0.378 | rtn1a | -0.021 |
| si:ch211-243a20.4 | 0.377 | si:ch211-288g17.3 | -0.021 |
| cd22 | 0.376 | sparc | -0.021 |
| sc:d0284 | 0.372 | tubb5 | -0.021 |
| fcer1g | 0.371 | her15.1 | -0.021 |
| acod1 | 0.369 | elavl3 | -0.020 |
| il10ra | 0.367 | h1f0 | -0.020 |
| il1b | 0.366 | rnasekb | -0.020 |
| spi1b | 0.366 | si:ch211-133n4.4 | -0.020 |
| cybb | 0.364 | si:ch73-46j18.5 | -0.020 |
| si:ch73-343l4.2 | 0.362 | stmn1a | -0.020 |
| CU499330.1 | 0.360 | cdh2 | -0.019 |
| c1qc | 0.355 | fam168a | -0.019 |
| lgals3bpb | 0.352 | gpm6ab | -0.019 |