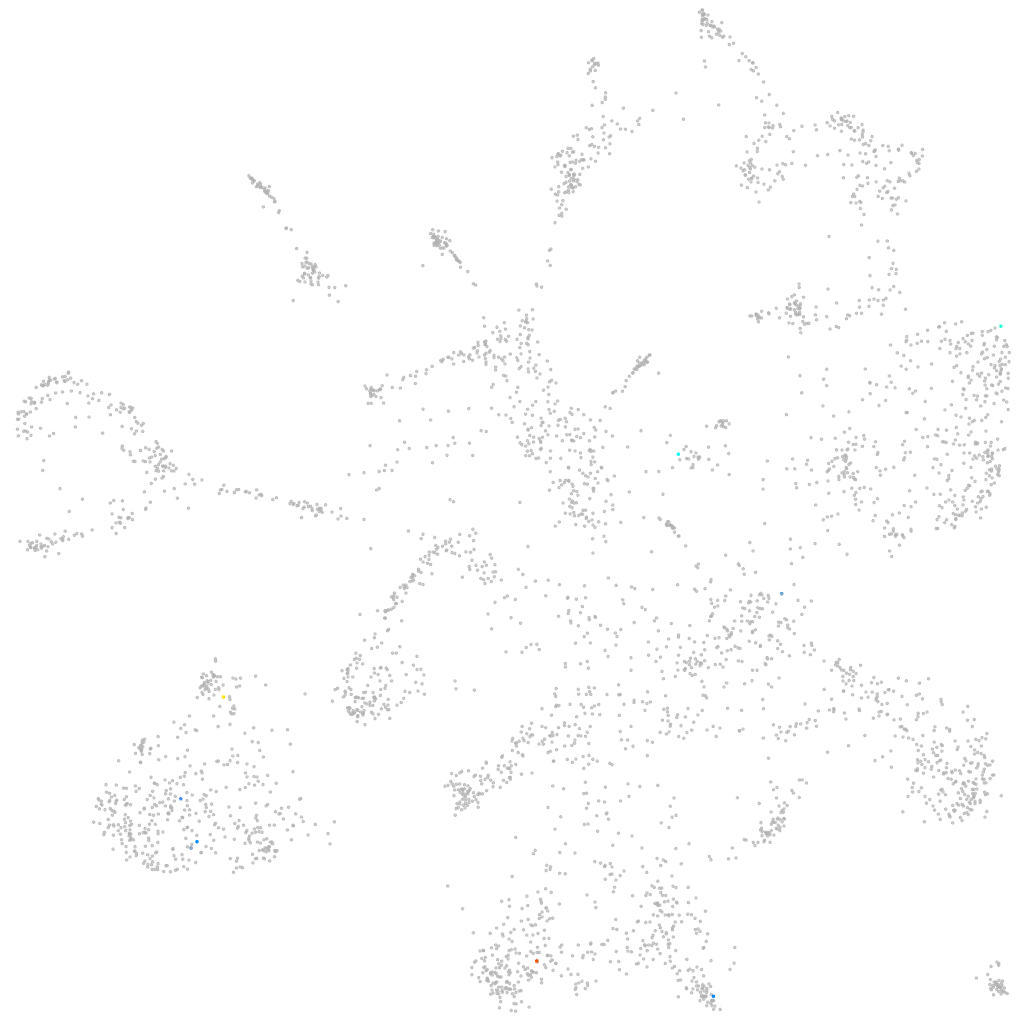
solute carrier family 2 member 6
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-160o17.6 | 0.615 | rps7 | -0.102 |
| dlg4a | 0.479 | rps19 | -0.100 |
| kcnj12a | 0.449 | rps10 | -0.092 |
| CABZ01057364.1 | 0.437 | rps2 | -0.088 |
| slc2a11a | 0.385 | rps14 | -0.088 |
| CT583728.12 | 0.356 | rps15a | -0.085 |
| sycn.1 | 0.330 | rps13 | -0.082 |
| cdk5r2b | 0.325 | rpl39 | -0.081 |
| CT583728.26 | 0.295 | rpl10a | -0.079 |
| pik3cg | 0.290 | rpl35a | -0.078 |
| limk1b | 0.287 | rpl12 | -0.078 |
| CABZ01073736.1 | 0.285 | rpl28 | -0.078 |
| tmem145 | 0.285 | rpl17 | -0.074 |
| gsg1l2b | 0.282 | rpl8 | -0.073 |
| CABZ01077121.1 | 0.280 | rpl9 | -0.071 |
| XLOC-011723 | 0.275 | slc25a5 | -0.070 |
| XLOC-036510 | 0.266 | rps25 | -0.068 |
| fut9b | 0.263 | rps4x | -0.067 |
| AL929576.1 | 0.258 | rplp0 | -0.067 |
| cnsta | 0.257 | rpl14 | -0.065 |
| fes | 0.257 | rpl22 | -0.064 |
| cdx4 | 0.255 | rpl31 | -0.063 |
| stxbp5l | 0.252 | rps15 | -0.063 |
| zgc:173577 | 0.251 | rps12 | -0.063 |
| sts | 0.250 | rps5 | -0.061 |
| p2ry6 | 0.241 | rpl18a | -0.060 |
| myo6b | 0.240 | zgc:114188 | -0.059 |
| c3a.1 | 0.237 | rack1 | -0.057 |
| nr0b1 | 0.234 | rpl36 | -0.056 |
| AL772300.1 | 0.233 | nme2b.1 | -0.056 |
| si:dkey-30e9.6 | 0.232 | rpl34 | -0.055 |
| fbxo31 | 0.230 | rps28 | -0.054 |
| FAM124A | 0.219 | rpl27 | -0.053 |
| frem2a | 0.215 | rps8a | -0.053 |
| crtac1b | 0.214 | ubc | -0.052 |