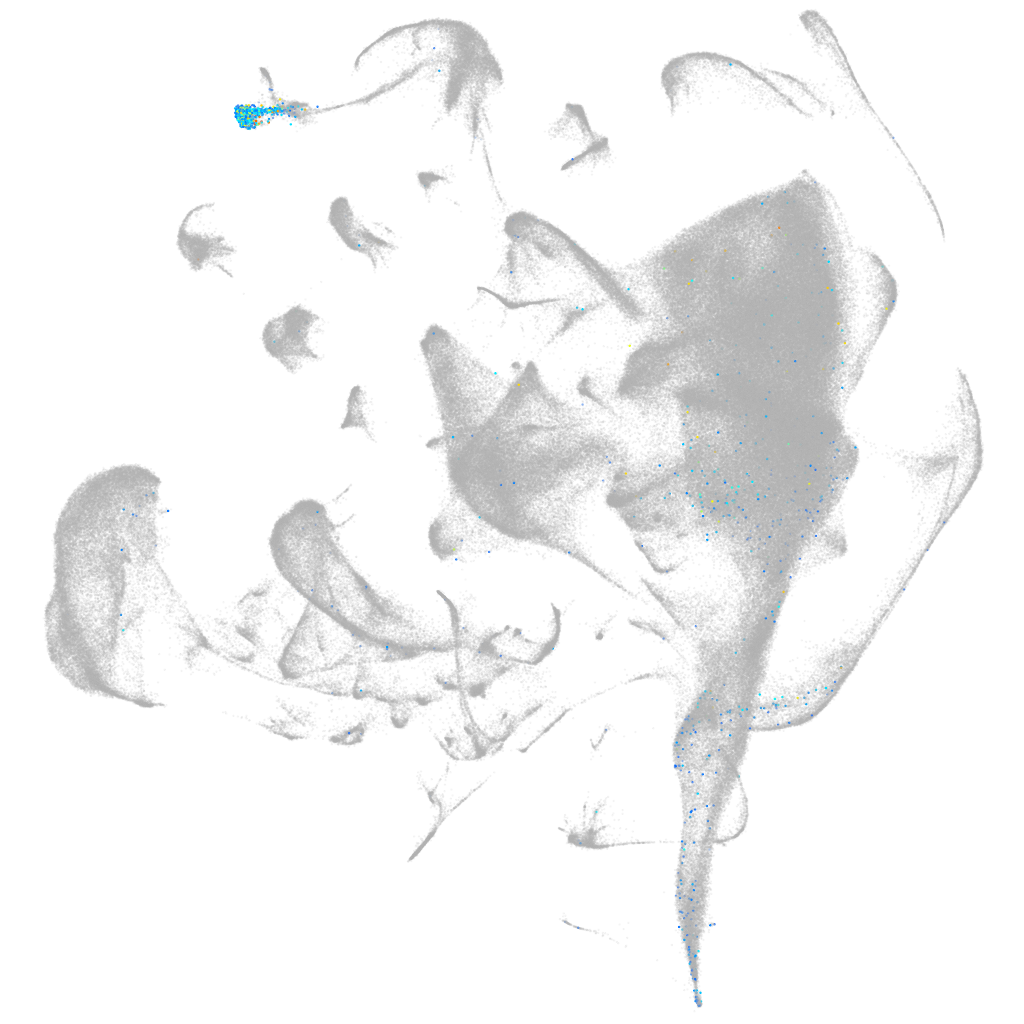
si:ch211-1o7.3
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm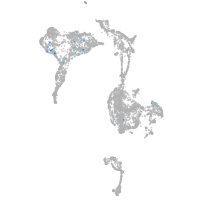 |
muscle 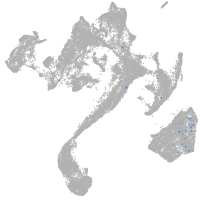 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 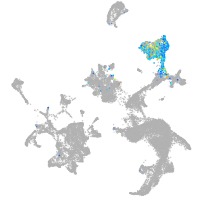 |
pronephros  |
neural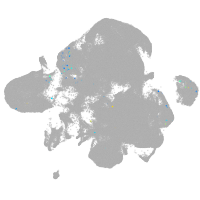 |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mpeg1.1 | 0.528 | mdka | -0.042 |
| havcr1 | 0.522 | fabp3 | -0.040 |
| cx32.2 | 0.519 | hmgb1b | -0.036 |
| cmklr1 | 0.501 | sparc | -0.036 |
| rnaset2l | 0.498 | hmgb3a | -0.034 |
| LOC108180725 | 0.492 | jpt1b | -0.033 |
| cxcr3.2 | 0.491 | foxp4 | -0.030 |
| si:dkey-5n18.1 | 0.487 | meis1b | -0.030 |
| mfap4 | 0.484 | pfn2l | -0.030 |
| spi1a | 0.481 | vdac1 | -0.030 |
| si:ch211-147m6.1 | 0.478 | pmp22a | -0.029 |
| si:ch211-194m7.3 | 0.478 | tuba1c | -0.029 |
| ccl34a.4 | 0.468 | mdkb | -0.027 |
| c1qb | 0.468 | nova2 | -0.027 |
| si:dkey-148a17.6 | 0.468 | tmeff1b | -0.027 |
| marco | 0.451 | tuba1a | -0.027 |
| CU499330.1 | 0.445 | gpm6aa | -0.026 |
| itgae.1 | 0.443 | si:ch211-137a8.4 | -0.026 |
| fcer1gl | 0.440 | tspan7 | -0.026 |
| spi1b | 0.427 | agrn | -0.025 |
| lgals3bpb | 0.405 | boc | -0.025 |
| c1qc | 0.395 | gpm6ab | -0.025 |
| slc2a6 | 0.390 | her6 | -0.025 |
| fcer1g | 0.385 | pbx4 | -0.025 |
| LOC101882264 | 0.385 | si:ch73-46j18.5 | -0.025 |
| irf8 | 0.384 | mfap2 | -0.025 |
| FO393424.2 | 0.383 | fabp7a | -0.024 |
| grna | 0.383 | mycb | -0.024 |
| laptm5 | 0.381 | rtn1a | -0.024 |
| FERMT3 (1 of many) | 0.378 | si:dkey-28b4.7 | -0.024 |
| zgc:173915 | 0.375 | dap1b | -0.024 |
| si:ch211-243a20.4 | 0.372 | her15.1 | -0.024 |
| adgrg3 | 0.370 | si:ch211-286o17.1 | -0.024 |
| sc:d0284 | 0.368 | col1a2 | -0.023 |
| ccl34b.1 | 0.367 | elavl3 | -0.023 |