solute carrier family 22 member 16
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 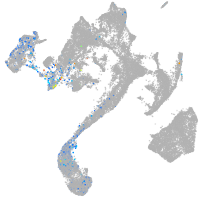 |
mural cells / non-skeletal muscle  |
mesenchyme 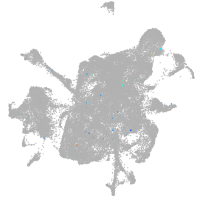 |
hematopoietic / vasculature 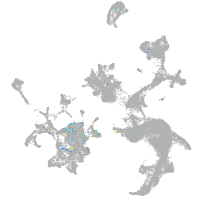 |
pronephros 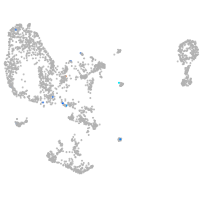 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mrc1a | 0.161 | hbae1.1 | -0.069 |
| hapln3 | 0.155 | hbae1.3 | -0.067 |
| dab2 | 0.151 | hbbe2 | -0.067 |
| lyve1b | 0.151 | prdx2 | -0.067 |
| cpn1 | 0.146 | hbae3 | -0.064 |
| stab2 | 0.145 | hbbe1.1 | -0.064 |
| gpr182 | 0.142 | hbbe1.3 | -0.063 |
| stab1 | 0.138 | cahz | -0.063 |
| c1qtnf6a | 0.138 | blvrb | -0.060 |
| hyal2b | 0.135 | hemgn | -0.058 |
| si:dkey-28n18.9 | 0.134 | hbbe1.2 | -0.056 |
| CU062454.1 | 0.133 | alas2 | -0.055 |
| ramp2 | 0.131 | tspo | -0.055 |
| flt4 | 0.124 | nt5c2l1 | -0.051 |
| plvapb | 0.122 | fth1a | -0.051 |
| CR677513.1 | 0.122 | gpx1a | -0.051 |
| lyve1a | 0.122 | slc4a1a | -0.051 |
| mtmr8 | 0.120 | si:ch211-250g4.3 | -0.050 |
| ccdc80 | 0.120 | epb41b | -0.050 |
| tspan4b | 0.120 | zgc:163057 | -0.050 |
| pcdh12 | 0.120 | creg1 | -0.048 |
| tgfbr2b | 0.120 | nmt1b | -0.046 |
| sypl2a | 0.120 | si:ch211-207c6.2 | -0.044 |
| myct1a | 0.120 | tmod4 | -0.041 |
| rab11bb | 0.118 | zgc:56095 | -0.040 |
| cd81a | 0.118 | tmem14ca | -0.040 |
| ctsla | 0.118 | znfl2a | -0.040 |
| cdh6 | 0.116 | sptb | -0.040 |
| si:ch211-145b13.6 | 0.116 | tfr1a | -0.039 |
| ckba | 0.116 | hbae5 | -0.039 |
| lamp2 | 0.116 | plac8l1 | -0.039 |
| lgmn | 0.115 | uros | -0.038 |
| si:ch211-66e2.3 | 0.115 | hdr | -0.038 |
| b3gnt7 | 0.115 | slc11a2 | -0.038 |
| LOC108182124 | 0.114 | klf1 | -0.037 |




