solute carrier family 14 member 2
ZFIN 
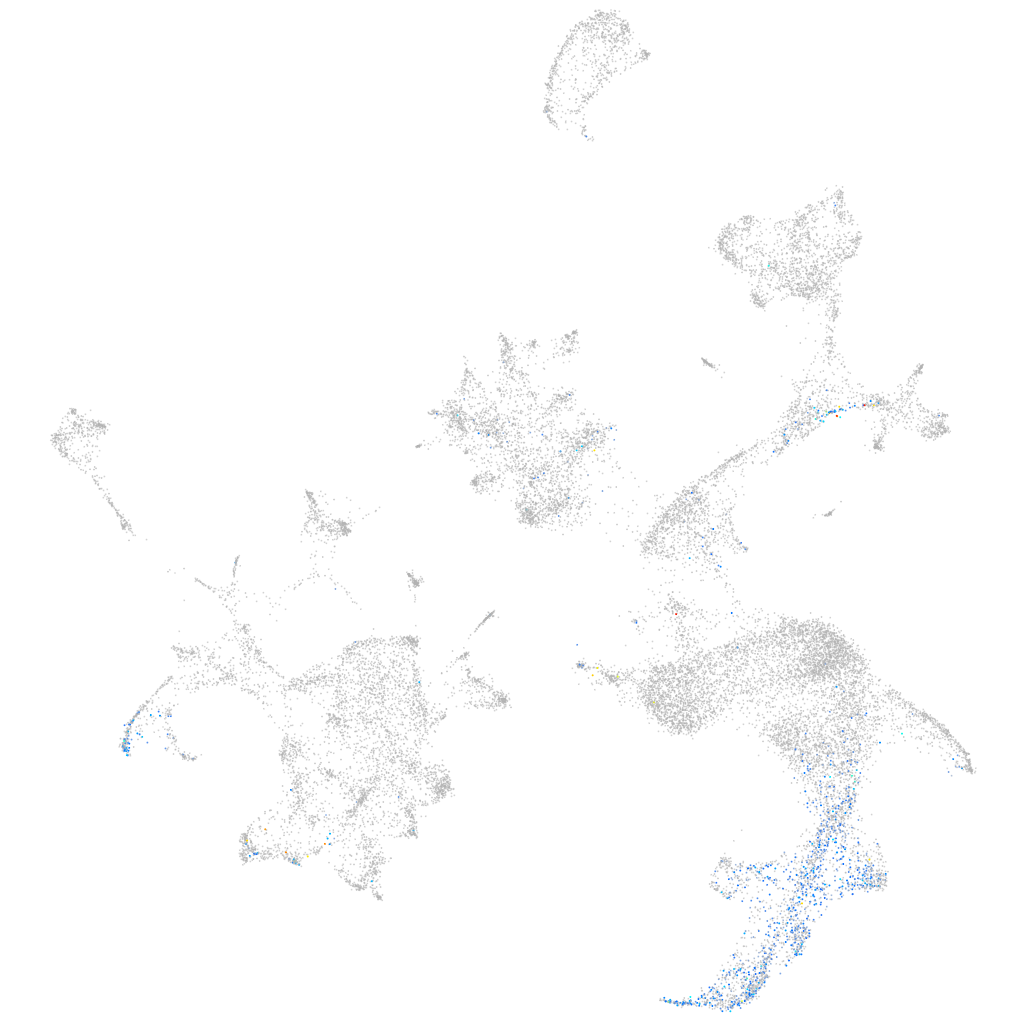
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hbbe3 | 0.265 | tmsb4x | -0.110 |
| hmbsb | 0.264 | marcksl1b | -0.108 |
| hmbsa | 0.263 | marcksl1a | -0.105 |
| cpox | 0.256 | actb2 | -0.092 |
| urod | 0.250 | ptmaa | -0.089 |
| alad | 0.224 | sec61g | -0.085 |
| znfl2a | 0.218 | ctsla | -0.085 |
| abcb10 | 0.203 | gapdhs | -0.081 |
| klf17 | 0.202 | ccni | -0.079 |
| cyp46a1.1 | 0.199 | cd63 | -0.077 |
| ppox | 0.194 | eif1b | -0.076 |
| tmem14ca | 0.194 | btg1 | -0.076 |
| fech | 0.192 | cd81a | -0.075 |
| blf | 0.180 | xbp1 | -0.075 |
| glrx5 | 0.177 | gpx1a | -0.075 |
| hdr | 0.174 | CR383676.1 | -0.074 |
| tfr1a | 0.172 | ppdpfb | -0.074 |
| si:dkey-261j4.4 | 0.171 | cotl1 | -0.073 |
| cldng | 0.168 | dap1b | -0.072 |
| si:dkey-261j4.3 | 0.167 | arpc4l | -0.072 |
| prdx3 | 0.165 | aldh9a1a.1 | -0.071 |
| slc10a4 | 0.162 | zgc:162730 | -0.071 |
| phlda2 | 0.160 | ckbb | -0.071 |
| mllt3 | 0.158 | itm2ba | -0.070 |
| zgc:163057 | 0.156 | fxyd6l | -0.070 |
| gata1a | 0.154 | arpc2 | -0.070 |
| slc25a37 | 0.153 | pnrc2 | -0.069 |
| slc11a2 | 0.153 | si:ch211-260e23.9 | -0.069 |
| anp32b | 0.152 | pvalb2 | -0.068 |
| uros | 0.151 | vat1 | -0.068 |
| txn | 0.149 | bloc1s6 | -0.067 |
| blvra | 0.149 | CT030188.1 | -0.067 |
| abce1 | 0.149 | hmgb3a | -0.066 |
| si:ch73-248e21.7 | 0.147 | akap12b | -0.066 |
| slc38a5a | 0.147 | ptmab | -0.066 |