"SLAIN motif family, member 1b"
ZFIN 
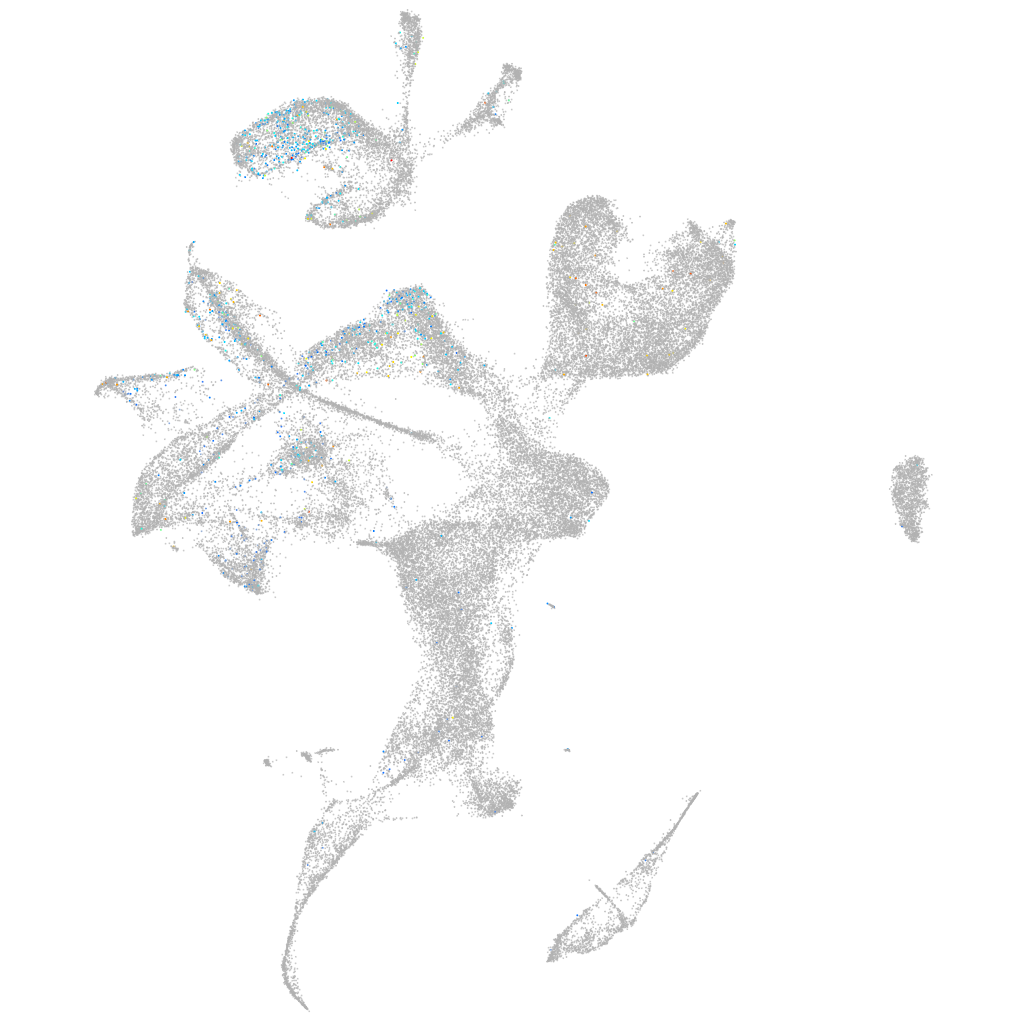
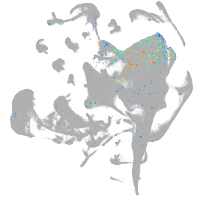
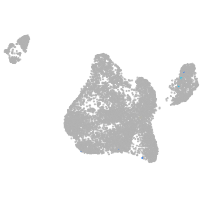
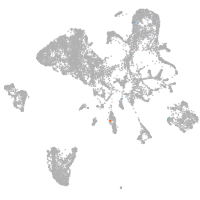
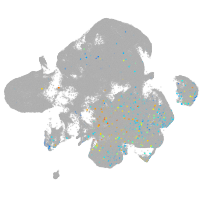
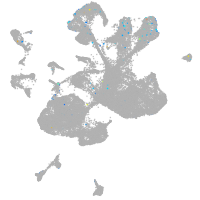
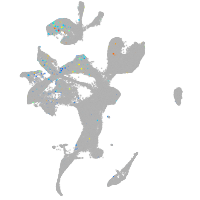
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sncb | 0.108 | hmgb2a | -0.069 |
| stxbp1a | 0.105 | mdka | -0.062 |
| stx1b | 0.103 | pcna | -0.062 |
| rtn1b | 0.102 | rrm1 | -0.059 |
| ywhag2 | 0.101 | her15.1 | -0.058 |
| olfm2a | 0.100 | ahcy | -0.056 |
| pax10 | 0.099 | COX7A2 (1 of many) | -0.055 |
| zgc:153426 | 0.099 | tspan7 | -0.054 |
| meis2b | 0.098 | msna | -0.053 |
| slc32a1 | 0.097 | banf1 | -0.052 |
| syt1a | 0.093 | ccnd1 | -0.052 |
| eno2 | 0.093 | mcm7 | -0.052 |
| zgc:65894 | 0.092 | id1 | -0.052 |
| tmsb2 | 0.091 | hes6 | -0.052 |
| si:ch73-290k24.5 | 0.091 | cldn5a | -0.051 |
| gng3 | 0.090 | nutf2l | -0.050 |
| LOC101886114 | 0.088 | lig1 | -0.050 |
| rbfox1 | 0.088 | stmn1a | -0.048 |
| elavl3 | 0.087 | tuba8l | -0.047 |
| gabrb4 | 0.087 | XLOC-003690 | -0.047 |
| sprn | 0.086 | dut | -0.047 |
| vamp2 | 0.085 | chaf1a | -0.047 |
| tfap2a | 0.085 | mcm2 | -0.046 |
| cacng3b | 0.083 | CABZ01005379.1 | -0.046 |
| plppr4b | 0.083 | rrm2 | -0.046 |
| pbx1a | 0.083 | fen1 | -0.045 |
| chgb | 0.081 | selenoh | -0.044 |
| basp1 | 0.081 | mcm6 | -0.044 |
| snap25a | 0.081 | ccna2 | -0.044 |
| myo10l1 | 0.081 | rpa2 | -0.044 |
| ppp1r14ba | 0.081 | mcm5 | -0.044 |
| calr | 0.080 | XLOC-003692 | -0.044 |
| dscama | 0.080 | cdca7b | -0.043 |
| cnrip1a | 0.080 | mcm4 | -0.043 |
| gnao1a | 0.080 | rpa3 | -0.043 |