si:dkey-261h17.1
ZFIN 
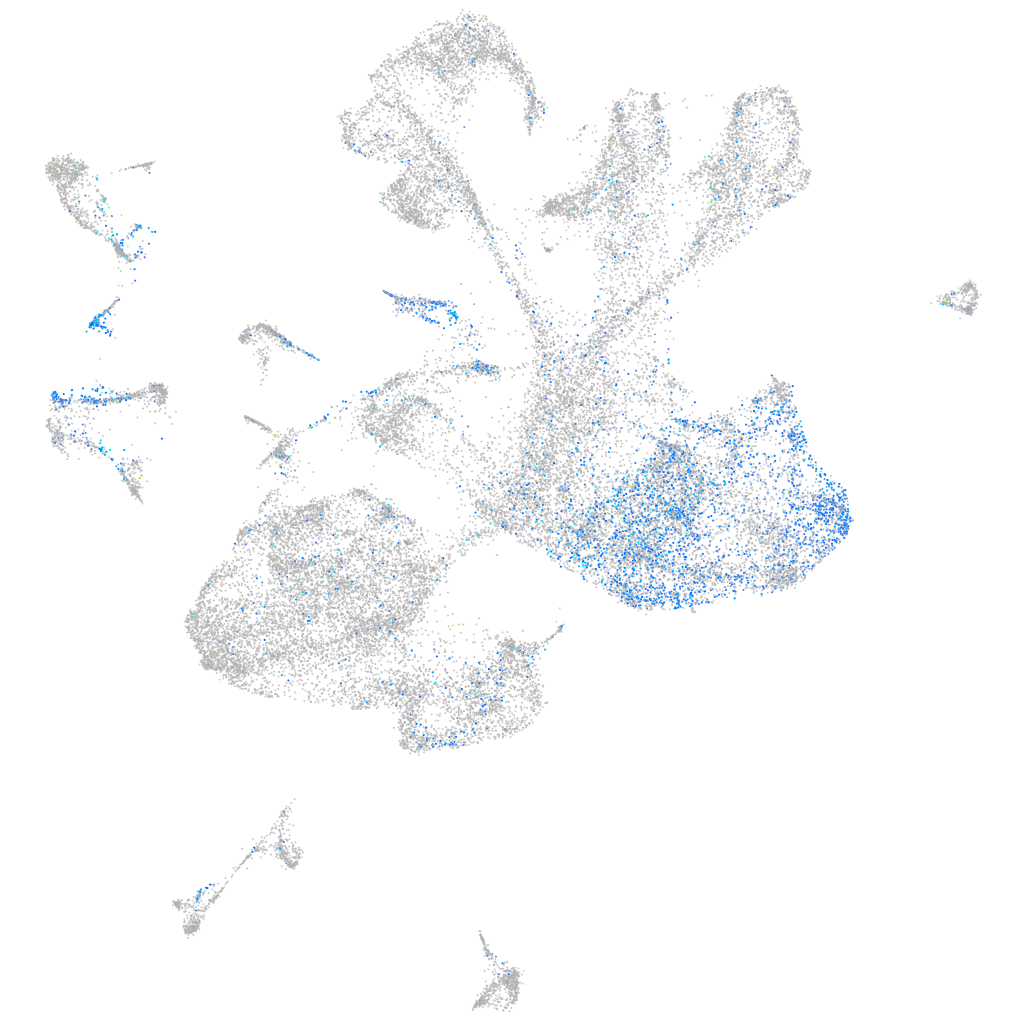
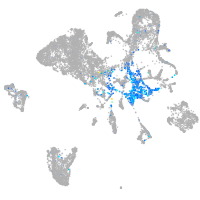
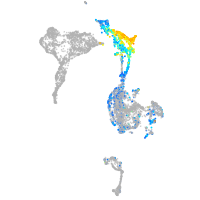
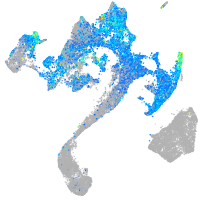
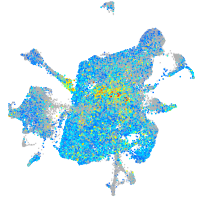
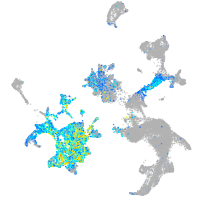
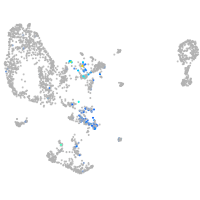
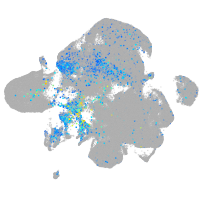
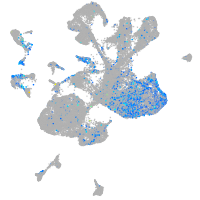
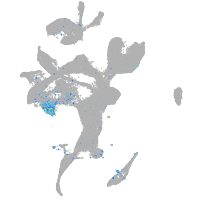
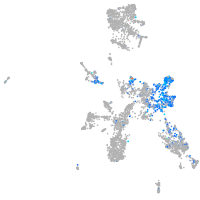
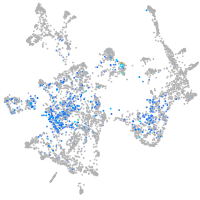
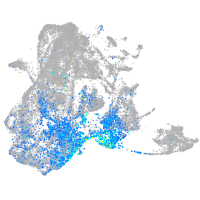
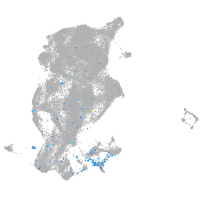
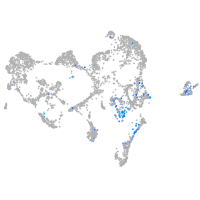
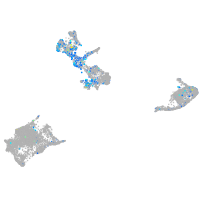
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cdx4 | 0.238 | ckbb | -0.190 |
| stmn1a | 0.231 | rtn1a | -0.150 |
| hmgb2a | 0.218 | gpm6aa | -0.147 |
| dut | 0.211 | slc1a2b | -0.130 |
| lbr | 0.200 | rnasekb | -0.130 |
| apela | 0.200 | CU467822.1 | -0.129 |
| chaf1a | 0.199 | gpm6ab | -0.129 |
| nradd | 0.198 | elavl3 | -0.123 |
| XLOC-042222 | 0.198 | gapdhs | -0.122 |
| pcna | 0.197 | fabp7a | -0.121 |
| mki67 | 0.196 | tmsb | -0.119 |
| cx43.4 | 0.195 | stmn1b | -0.118 |
| anp32b | 0.191 | CR383676.1 | -0.117 |
| znfl2a | 0.190 | fez1 | -0.115 |
| dek | 0.190 | myt1b | -0.112 |
| rrm1 | 0.188 | elavl4 | -0.110 |
| cks1b | 0.188 | si:dkeyp-75h12.5 | -0.107 |
| banf1 | 0.186 | gnao1a | -0.107 |
| lamb1a | 0.186 | ywhah | -0.107 |
| tuba8l4 | 0.185 | gng3 | -0.107 |
| XLOC-001964 | 0.185 | CR848047.1 | -0.104 |
| hmga1a | 0.184 | cotl1 | -0.102 |
| ccna2 | 0.184 | nova2 | -0.101 |
| si:ch73-281n10.2 | 0.180 | stxbp1a | -0.100 |
| hmgb2b | 0.180 | pvalb1 | -0.100 |
| sp5l | 0.179 | atp6v0cb | -0.100 |
| zgc:110216 | 0.179 | atp6v1e1b | -0.099 |
| nasp | 0.177 | gng2 | -0.099 |
| smc2 | 0.174 | sncb | -0.098 |
| fen1 | 0.173 | dpysl2b | -0.098 |
| zic2b | 0.173 | dpysl3 | -0.098 |
| nutf2l | 0.172 | acbd7 | -0.098 |
| wnt11r | 0.172 | vamp2 | -0.097 |
| rrm2 | 0.171 | pvalb2 | -0.097 |
| BX005254.3 | 0.170 | stx1b | -0.096 |