si:dkey-230p4.1
ZFIN 
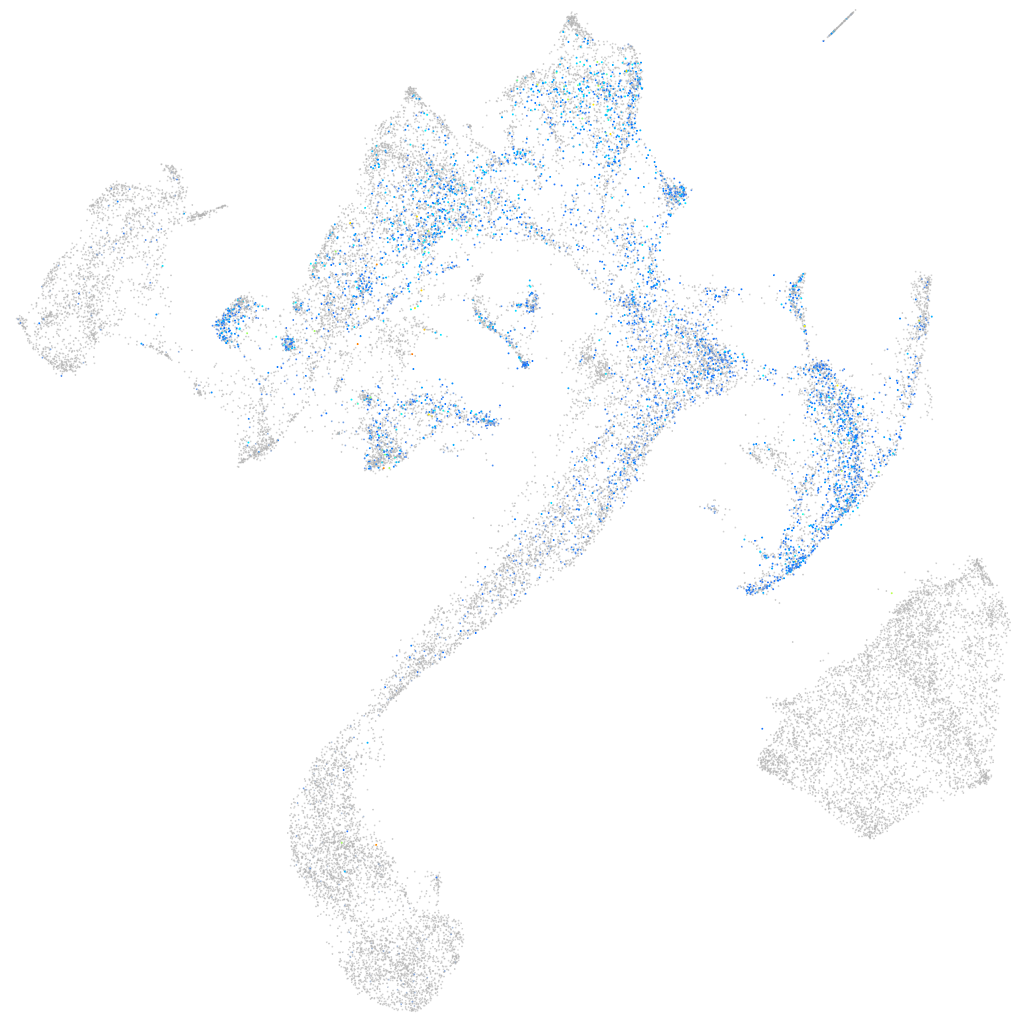
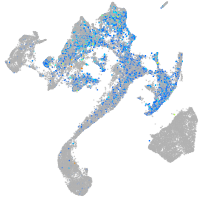
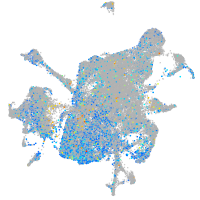
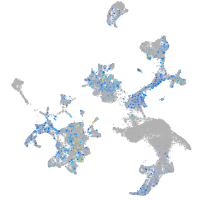
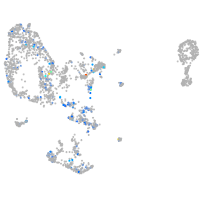
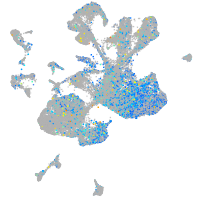
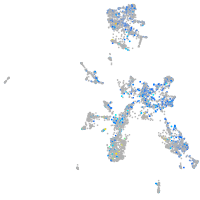
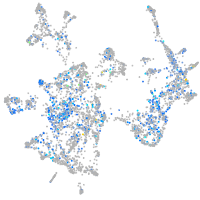
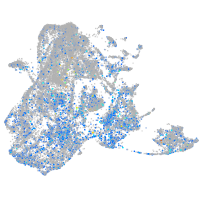
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| h3f3a | 0.327 | gapdh | -0.185 |
| hnrnpa0l | 0.284 | aldoab | -0.177 |
| zgc:153409 | 0.277 | ak1 | -0.176 |
| dut | 0.271 | actc1b | -0.176 |
| zgc:165555.3 | 0.265 | tnnc2 | -0.174 |
| zgc:56493 | 0.263 | ckmb | -0.173 |
| tubb4b | 0.259 | ckma | -0.172 |
| tuba8l | 0.249 | atp2a1 | -0.171 |
| sinhcafl | 0.245 | tmem38a | -0.169 |
| hmgn2 | 0.238 | neb | -0.169 |
| cks2 | 0.231 | acta1b | -0.166 |
| si:dkey-261m9.17 | 0.230 | srl | -0.162 |
| ewsr1b | 0.223 | actn3a | -0.160 |
| cenpe | 0.220 | ldb3b | -0.160 |
| cfl1 | 0.216 | ldb3a | -0.159 |
| si:dkey-261h17.1 | 0.216 | actn3b | -0.159 |
| aspm | 0.215 | tpma | -0.159 |
| tubb2b | 0.213 | si:ch73-367p23.2 | -0.158 |
| mfap2 | 0.213 | cav3 | -0.158 |
| ppiab | 0.212 | mybphb | -0.156 |
| tuba1b | 0.211 | CABZ01078594.1 | -0.155 |
| actb1 | 0.210 | desma | -0.154 |
| si:ch1073-429i10.3.1 | 0.210 | myom1a | -0.154 |
| COX7A2 (1 of many) | 0.209 | tmod4 | -0.153 |
| hsp90ab1 | 0.207 | smyd1a | -0.152 |
| ppib | 0.207 | mylpfa | -0.152 |
| tuba8l4 | 0.205 | ank1a | -0.152 |
| marcksl1a | 0.205 | pgam2 | -0.152 |
| sall4 | 0.204 | casq2 | -0.152 |
| CABZ01058261.1 | 0.203 | nme2b.2 | -0.151 |
| tacc3 | 0.203 | si:ch211-266g18.10 | -0.151 |
| ptmaa | 0.203 | tnnt3a | -0.150 |
| cks1b | 0.200 | hhatla | -0.149 |
| cdk1 | 0.199 | mylz3 | -0.149 |
| rrm2 | 0.198 | ttn.2 | -0.147 |